3UZB
 
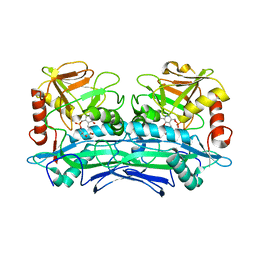 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UZO
 
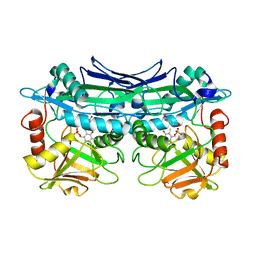 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
4R8V
 
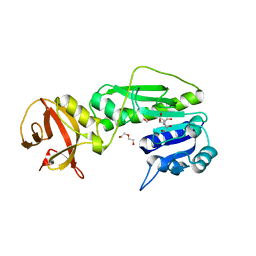 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with formate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-09-03 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3V1Y
 
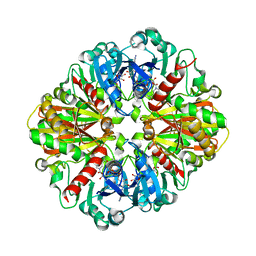 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
4QPD
 
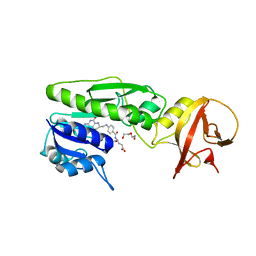 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 10-formyltetrahydrofolate dehydrogenase, DI(HYDROXYETHYL)ETHER | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QPC
 
 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (Y200A) from zebrafish | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3UYY
 
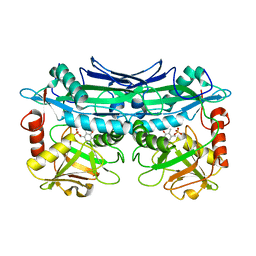 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
4TT8
 
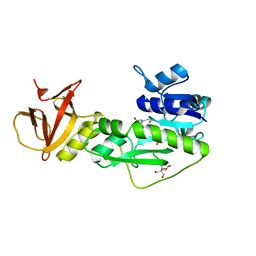 | | Crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase (wild-type) complex with 10-formyl-5,8-dideazafolate | | Descriptor: | 10-formyltetrahydrofolate dehydrogenase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(4-{[(2-amino-4-hydroxyquinazolin-6-yl)methyl](formyl)amino}benzoyl)-L-glutamic acid | | Authors: | Lin, C.C, Chen, C.J, Fu, T.F, Chuankhayan, P, Kao, T.T, Chang, W.N. | | Deposit date: | 2014-06-20 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures of the hydrolase domain of zebrafish 10-formyltetrahydrofolate dehydrogenase and its complexes reveal a complete set of key residues for hydrolysis and product inhibition.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XTH
 
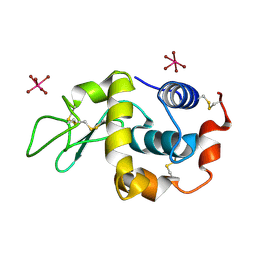 | | K2PtBr6 binding to lysozyme | | Descriptor: | HEXABROMOPLATINATE(IV), LYSOZYME C | | Authors: | Helliwell, J.R, Bell, A.M.T, Bryant, P, Fisher, S, Habash, J, Helliwell, M, Margiolaki, I, Kaenket, S, Watier, Y, Wright, J, Yalamanchili, S.K. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Dependent Analysis of K2Ptbr6 Binding to Lysozyme Studied by Protein Powder and Single Crystal X-Ray Analysis
Z.Kristallogr., 225, 2010
|
|
2FS1
 
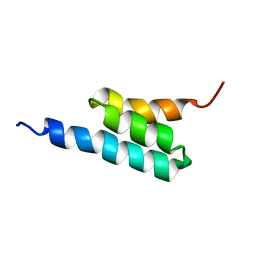 | | solution structure of PSD-1 | | Descriptor: | PSD-1 | | Authors: | He, Y, Rozak, D.A, Sari, N, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2006-01-20 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity.
Biochemistry, 45, 2006
|
|
2KJU
 
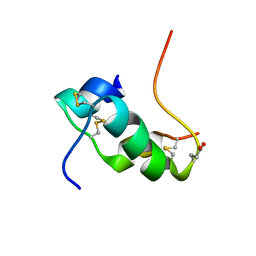 | | NMR structure of human insulin mutant glu-b21-d-glu, his-b10 asp pro-b28-lys, lys-b29-pro, 20 structures | | Descriptor: | Insulin | | Authors: | Hua, Q.X, Huang, K, Hu, S.Q, Katsoyanni, P, Weiss, M.A. | | Deposit date: | 2009-06-10 | | Release date: | 2010-06-16 | | Last modified: | 2021-10-13 | | Method: | SOLUTION NMR | | Cite: | Acceleration of Protein Fibrillation by Chiral Destabilization of Beta-Turn
To be Published
|
|
4DNW
 
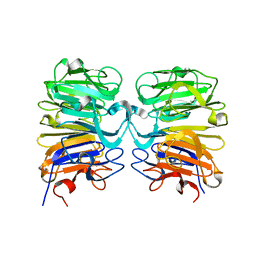 | | Crystal structure of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNV
 
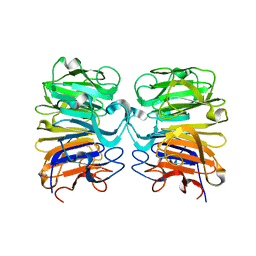 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
3O0I
 
 | |
3O2F
 
 | |
5B78
 
 | |
5B75
 
 | |
5B76
 
 | |
5B77
 
 | |
5B79
 
 | | Crystal structure of DPF2 double PHD finger | | Descriptor: | ZINC ION, Zinc finger protein ubi-d4 | | Authors: | Li, H, Xiong, X. | | Deposit date: | 2016-06-05 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2
Nat.Chem.Biol., 12, 2016
|
|
8JAY
 
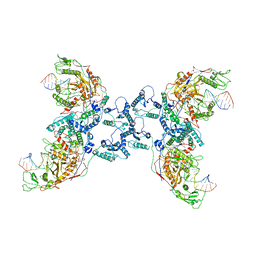 | | CrtSPARTA Octamer bound with guide-target | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Guo, L.J, Huang, P.P, Li, Z.X, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
7XZO
 
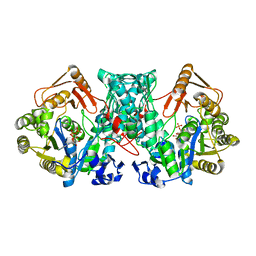 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
7XZN
 
 | |
7XZP
 
 | |
