4D26
 
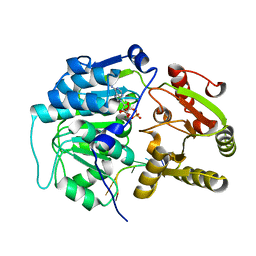 | | Crystal structure of the Bombyx mori Vasa helicase (E339Q) in complex with RNA,ADP and Pi | | Descriptor: | 5'-R(*UP*GP*AP*CP*AP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, BMVLG PROTEIN, ... | | Authors: | Spinelli, P, Pillai, R.S, Kadlec, J, Cusack, S. | | Deposit date: | 2014-05-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RNA Clamping by Vasa Assembles a Pirna Amplifier Complex on Transposon Transcripts.
Cell(Cambridge,Mass.), 157, 2014
|
|
6Y6K
 
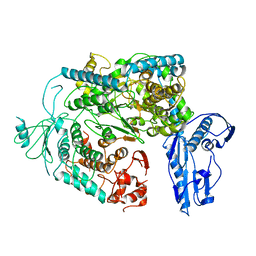 | | Cryo-EM structure of a Phenuiviridae L protein | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase | | Authors: | Vogel, D, Thorkelsson, S.R, Quemin, E, Meier, K, Kouba, T, Gogrefe, N, Busch, C, Reindl, S, Guenther, S, Cusack, S, Gruenewald, K, Rosenthal, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
1E6J
 
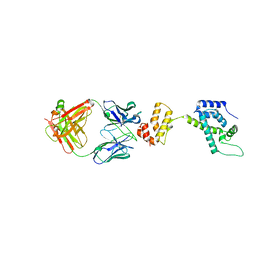 | | Crystal structure of HIV-1 capsid protein (p24) in complex with Fab13B5 | | Descriptor: | CAPSID PROTEIN P24, IMMUNOGLOBULIN | | Authors: | Berthet-Colominas, C, Monaco, S, Novelli, A, Sibai, G, Mallet, F, Cusack, S. | | Deposit date: | 2000-08-18 | | Release date: | 2000-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mutual Conformational Adaptations in Antigen and Antibody Upon Complex Formation between an Fab and HIV-1 Capsid Protein P24
Structure, 8, 2000
|
|
1NSB
 
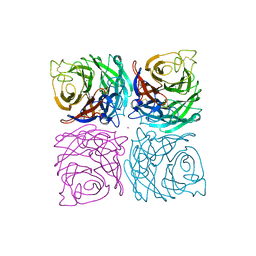 | |
5MUS
 
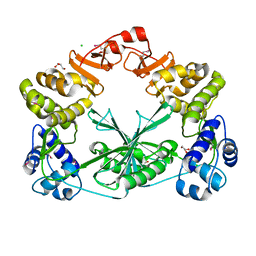 | | Structure of the C-terminal domain of a reptarenavirus L protein | | Descriptor: | CHLORIDE ION, GLYCEROL, L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
2VDW
 
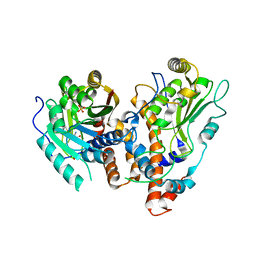 | | Guanosine N7 methyl-transferase sub-complex (D1-D12) of the vaccinia virus mRNA capping enzyme | | Descriptor: | MRNA-CAPPING ENZYME SMALL SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | De la Pena, M, Kyrieleis, O.J.P, Cusack, S. | | Deposit date: | 2007-10-12 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into the Mechanism and Evolution of the Vaccinia Virus Mrna CAP N7 Methyl- Transferase.
Embo J., 26, 2007
|
|
2V70
 
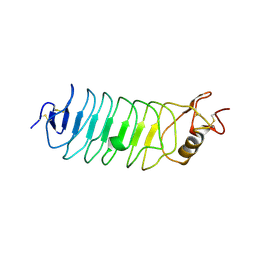 | | Third LRR domain of human Slit2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SLIT HOMOLOG 2 PROTEIN N-PRODUCT | | Authors: | Morlot, C, Cusack, S, McCarthy, A.A. | | Deposit date: | 2007-07-24 | | Release date: | 2007-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Production of Slit2 Lrr Domains in Mammalian Cells for Structural Studies and the Structure of Human Slit2 Domain 3.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4WSA
 
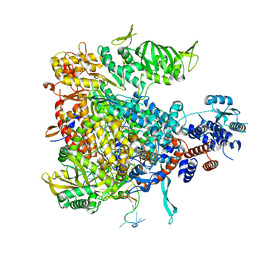 | | Crystal structure of Influenza B polymerase bound to the vRNA promoter (FluB1 form) | | Descriptor: | Influenza B vRNA promoter 3' end, Influenza B vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
1914
 
 | | SIGNAL RECOGNITION PARTICLE ALU RNA BINDING HETERODIMER, SRP9/14 | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHATE ION, SIGNAL RECOGNITION PARTICLE 9/14 FUSION PROTEIN | | Authors: | Birse, D, Kapp, U, Strub, K, Cusack, S, Aberg, A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
EMBO J., 16, 1997
|
|
2V9Q
 
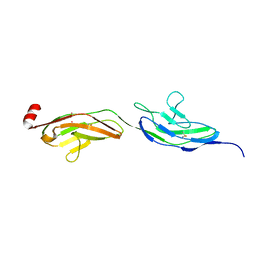 | |
5EPI
 
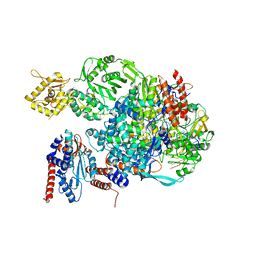 | |
5M3J
 
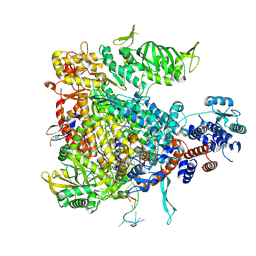 | | Influenza B polymerase bound to four heptad repeats of serine 5 phosphorylated Pol II CTD | | Descriptor: | DNA-directed RNA polymerase subunit, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Lukarska, M, Pflug, A, Cusack, S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of an essential interaction between influenza polymerase and Pol II CTD.
Nature, 541, 2017
|
|
5M3H
 
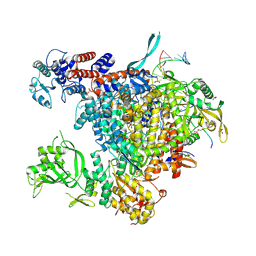 | | Bat influenza A/H17N10 polymerase bound to four heptad repeats of serine 5 phosphorylated Pol II CTD | | Descriptor: | PHOSPHATE ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Lukarska, M, Pflug, A, Cusack, S. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of an essential interaction between influenza polymerase and Pol II CTD.
Nature, 541, 2017
|
|
5FMZ
 
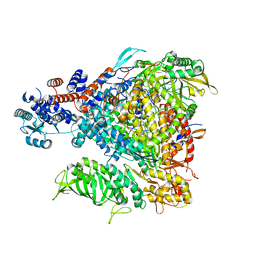 | | Crystal structure of Influenza B polymerase with bound 5' vRNA | | Descriptor: | 5'-R(*AP*GP*UP*AP*GP*UP*AP*AP*CP*AP*AP*GP)-3', POLYMERASE ACIDIC PROTEIN, POLYMERASE BASIC PROTEIN 2, ... | | Authors: | Guilligay, D, Cusack, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
8AZA
 
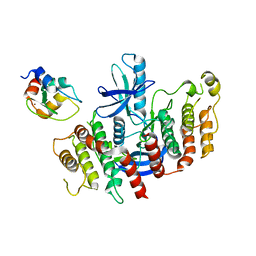 | | Structure of RIP2K dimer bound to the XIAP BIR2 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, Receptor-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2022-09-05 | | Release date: | 2022-10-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure shows that the BIR2 domain of E3 ligase XIAP binds across the RIPK2 kinase dimer interface.
Life Sci Alliance, 6, 2023
|
|
8ASG
 
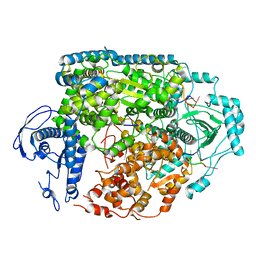 | | Structure of the SFTSV L protein bound in a resting state [RESTING] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*AP*UP*GP*A*)-3'), RNA (5'-R(*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U*)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8BY6
 
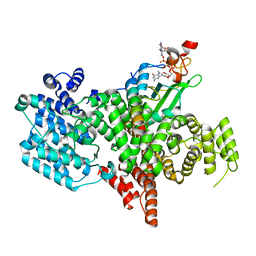 | | Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
8CI5
 
 | | Structure of the SNV L protein bound to 5' RNA | | Descriptor: | RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*AP*CP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Meier, K, Thorkelsson, S.R, Durieux Trouilleton, Q, Vogel, D, Yu, D, Kosinski, J, Cusack, S, Malet, H, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of the Sin Nombre virus L protein.
Plos Pathog., 19, 2023
|
|
1SRY
 
 | |
6Z6G
 
 | | Cryo-EM structure of La Crosse virus polymerase at pre-initiation stage | | Descriptor: | 3'vRNA 1-16, 5'vRNA 1-10, 5'vRNA 9-16, ... | | Authors: | Arragain, B, Effantin, G, Gerlach, P, Reguera, J, Schoehn, G, Cusack, S, Malet, H. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
4CGX
 
 | | Crystal structure of the N-terminal domain of the PA subunit of Thogoto virus polymerase (form 1) | | Descriptor: | POLYMERASE ACIDIC PROTEIN | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
4CY1
 
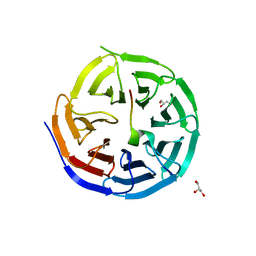 | | Crystal structure of the KANSL1-WDR5 complex. | | Descriptor: | GLYCEROL, KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CY2
 
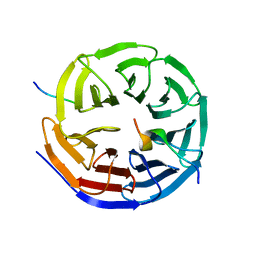 | | Crystal structure of the KANSL1-WDR5-KANSL2 complex. | | Descriptor: | KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, KAT8 REGULATORY NSL COMPLEX SUBUNIT 2, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CY3
 
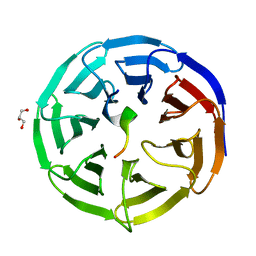 | | Crystal structure of the NSL1-WDS complex. | | Descriptor: | CG4699, ISOFORM D, GLYCEROL, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CY5
 
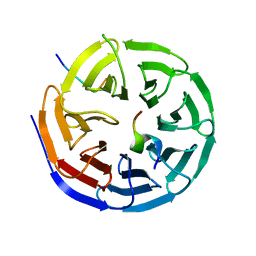 | | Crystal structure of the NSL1-WDS-NSL2 complex. | | Descriptor: | CG4699, ISOFORM D, DIM GAMMA-TUBULIN 1, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
