6P46
 
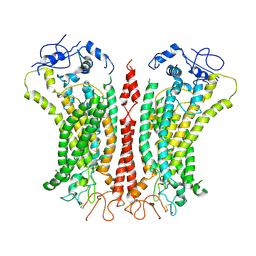 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P47
 
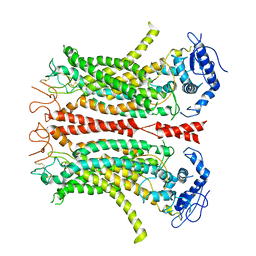 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | Descriptor: | Anoctamin-6 | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P48
 
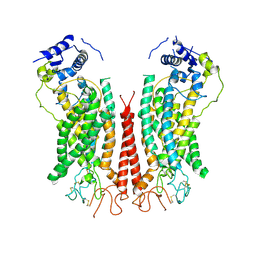 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | Deposit date: | 2019-05-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
5VKQ
 
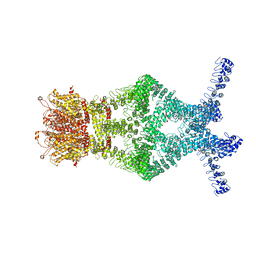 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | Authors: | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | Deposit date: | 2017-04-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
6O20
 
 | | Cryo-EM structure of TRPV5 with calmodulin bound | | Descriptor: | CALCIUM ION, Calmodulin, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O1N
 
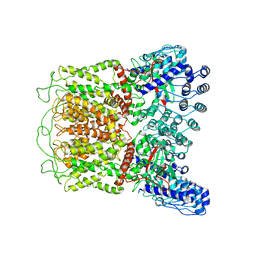 | | Cryo-EM structure of TRPV5 (1-660) in nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3IYV
 
 | | Clathrin D6 coat as full-length Triskelions | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Johnson, G.T, Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy.
Nature, 432, 2004
|
|
6O1U
 
 | | Cryo-EM structure of TRPV5 W583A in nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O1P
 
 | | Cryo-EM structure of full length TRPV5 in nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3J9I
 
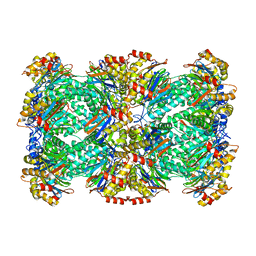 | | Thermoplasma acidophilum 20S proteasome | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Li, X, Mooney, P, Zheng, S, Booth, C, Braunfeld, M.B, Gubbens, S, Agard, D.A, Cheng, Y. | | Deposit date: | 2015-02-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM.
Nat.Methods, 10, 2013
|
|
8T0C
 
 | |
8T3L
 
 | | TRPV1 in nanodisc bound with 2 LPA molecules in neighboring monomers | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8T0E
 
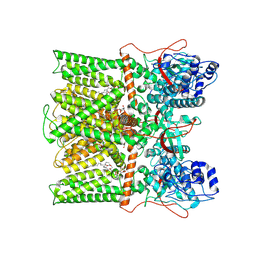 | | TRPV1 in Nanodisc not bound with lysophosphatidic acid (apo) | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Cheng, Y. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8T10
 
 | | TRPV1 in nanodisc bound with two LPA molecules in opposite monomers | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8T3M
 
 | | TRPV1 in nanodisc bound with 3 LPA molecules | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8T0Y
 
 | | TRPV1 in nanodisc bound with one LPA in one monomer | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, ... | | Authors: | Arnold, W.R, Cheng, Y. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3L
 
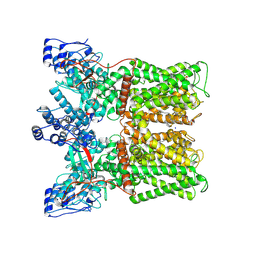 | | TRPV1 in nanodisc bound with empty vanilloid binding pocket at 25C | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U30
 
 | | TRPV1 in nanodisc bound with diC8-PIP2 in the closed state | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3C
 
 | | TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 2 (monomer) | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U2Z
 
 | | TRPV1 in nanodisc bound with diC8-PIP2 in the dilated state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3J
 
 | | TRPV1 in nanodisc bound with empty vanilloid binding pocket at 4C | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U4D
 
 | | TRPV1 in nanodisc bound with PI-Br4, consensus structure | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3A
 
 | | TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 1 (monomer) | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U43
 
 | | TRPV1 in nanodisc bound with PIP2-Br4 | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6V
 
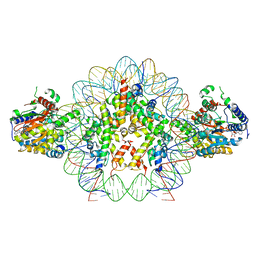 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
