5D5F
 
 | | In meso in situ serial X-ray crystallography structure of lysozyme by bromine-SAD at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D58
 
 | | In meso in situ serial X-ray crystallography structure of the PepTSt-Ala-Phe complex at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALANINE, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5D
 
 | | In meso in situ serial X-ray crystallography structure of AlgE at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Ma, P, Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D7G
 
 | |
5ZF2
 
 | | Crystal structure of Trxlp from Edwardsiella tarda EIB202 | | Descriptor: | SULFATE ION, Thioredoxin (H-type,TRX-H) | | Authors: | Yang, C, Quan, S. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Edwardsiella piscicida thioredoxin-like protein inhibits ASK1-MAPKs signaling cascades to promote pathogenesis during infection.
Plos Pathog., 15, 2019
|
|
6AX7
 
 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
5TH9
 
 | | Structure determination of a potent, selective antibody inhibitor of human MMP9 (GS-5745 bound to MMP-9) | | Descriptor: | CALCIUM ION, COBALT HEXAMMINE(III), GS-5745 Fab heavy chain, ... | | Authors: | Appleby, T.C, Greenstein, A.E, Kwon, H.J. | | Deposit date: | 2016-09-29 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Biochemical characterization and structure determination of a potent, selective antibody inhibitor of human MMP9.
J. Biol. Chem., 292, 2017
|
|
5D53
 
 | | In meso in situ serial X-ray crystallography structure of insulin at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D56
 
 | | In meso in situ serial X-ray crystallography structure of diacylglycerol kinase, DgkA, at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Howe, N, Olieric, V, Warshamanage, R, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D6L
 
 | | beta2AR-T4L - CIM | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Ma, P, Caffrey, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
5E2U
 
 | |
4IF2
 
 | |
2MU3
 
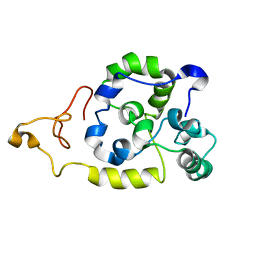 | | Spider wrapping silk fibre architecture arising from its modular soluble protein precursor | | Descriptor: | Aciniform spidroin 1 | | Authors: | Xu, L, Tremblay, M, Meng, Q, Liu, X, Rainey, J.K, Lefevre, T, Sarker, M, Orrell, K.E, Leclerc, J, Pezolet, M, Auger, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spider wrapping silk fibre architecture arising from its modular soluble protein precursor.
Sci Rep, 5, 2015
|
|
3RFU
 
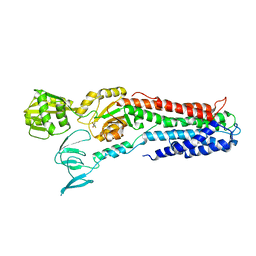 | | Crystal structure of a copper-transporting PIB-type ATPase | | Descriptor: | Copper efflux ATPase, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Gourdon, P, Liu, X, Skjorringe, T, Morth, J.P, Birk Moller, L, Panyella Pedersen, B, Nissen, P. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a copper-transporting PIB-type ATPase.
Nature, 475, 2011
|
|
2B96
 
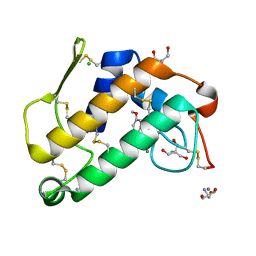 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | Authors: | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8HGR
 
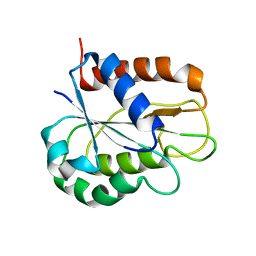 | | The apo-flavodoxin monomer from Synechococcus elongatus PCC 7942 | | Descriptor: | CHLORIDE ION, Flavodoxin, MAGNESIUM ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
8HGQ
 
 | | The apo-flavodoxin dimer from Synechococcus elongatus PCC 7942 | | Descriptor: | Flavodoxin, PHOSPHATE ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
7D72
 
 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D73
 
 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D74
 
 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (state II) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, Mannose-1-phosphate guanyltransferase beta | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-19 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8U47
 
 | |
8U46
 
 | |
8U48
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 D161A-E163A inactive Endoglycosidase in complex with high-mannose N-glycan (Man9GlcNAc2) substrate | | Descriptor: | Endo-beta-N-acetylglucosaminidase, PHOSPHATE ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sastre, D.E, Sultana, N, Navarro, M.V.A.S, Sundberg, E.J. | | Deposit date: | 2023-09-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
8U9F
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 in complex with NaI | | Descriptor: | 1,2-ETHANEDIOL, Endo-beta-N-acetylglucosaminidase, IODIDE ION, ... | | Authors: | Sastre, D.E, Navarro, M.V.A.S, Sundberg, E.J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
