7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5EK9
 
 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline inhibitor | | Descriptor: | Bromodomain-containing protein 2, propan-2-yl ~{N}-[(2~{S},4~{R})-1-ethanoyl-6-(furan-2-yl)-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Picaud, S, Fedorov, O, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploiting a water network to achieve enthalpy-driven, bromodomain-selective BET inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
3OP5
 
 | | Human vaccinia-related kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase VRK1, ... | | Authors: | Allerston, C.K, Uttarkar, S, Savitsky, P, Elkins, J.M, Filippakopoulos, P, Krojer, T, Rellos, P, Fedorov, O, Eswaran, J, Brenner, B, Keates, T, Das, S, King, O, Chalk, R, Berridge, G, von Delft, F, Gileadi, O, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
3H9F
 
 | | Crystal Structure of Human Dual Specificity Protein Kinase (TTK) in complex with a pyrimido-diazepin ligand | | Descriptor: | 9-cyclopentyl-2-(4-(4-hydroxypiperidin-1-yl)-2-methoxyphenylamino)-5-methyl-8,9-dihydro-5H-pyrimido[4,5-b][1,4]diazepin -6(7H)-one, Dual specificity protein kinase TTK, MAGNESIUM ION | | Authors: | Filippakopoulos, P, Soundararajan, M, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, Yue, W, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Kwiatkowski, N, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
3GFW
 
 | | Crystal Structure of Human Dual Specificity Protein Kinase (TTK) in complex with a pyrolo-pyridin ligand | | Descriptor: | 1-(4-(4-(2-(isopropylsulfonyl)phenylamino)-1H-pyrrolo[2,3-b]pyridin-6-ylamino)-3-methoxyphenyl)piperidin-4-ol, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Choi, H, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
3Q4U
 
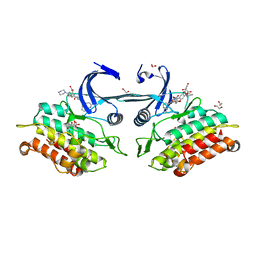 | | Crystal structure of the ACVR1 kinase domain in complex with LDN-193189 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C.D.O, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C.K, Krojer, T, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-24 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
3H9R
 
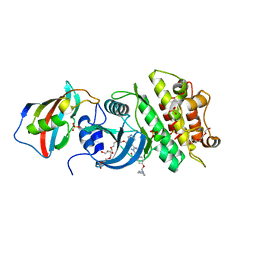 | | Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin | | Descriptor: | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Activin receptor type-1, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Chaikuad, A, Alfano, I, Shrestha, B, Muniz, J.R.C, Petrie, K, Fedorov, O, Phillips, C, Bishop, S, Mahajan, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Bone Morphogenetic Protein Receptor ALK2 and Implications for Fibrodysplasia Ossificans Progressiva.
J.Biol.Chem., 287, 2012
|
|
3IQ7
 
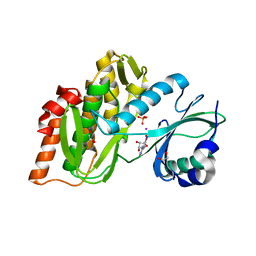 | | Crystal Structure of human Haspin in complex with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-19 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and functional characterization of the atypical human kinase haspin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G2F
 
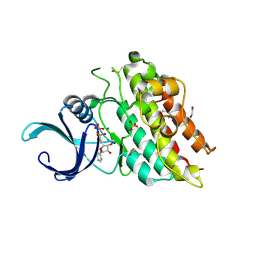 | | Crystal structure of the kinase domain of bone morphogenetic protein receptor type II (BMPR2) at 2.35 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bone morphogenetic protein receptor type-2, ... | | Authors: | Chaikuad, A, Thangaratnarajah, C, Roos, A.K, Filippakopoulos, P, Salah, E, Phillips, C, Keates, T, Fedorov, O, Chalk, R, Petrie, K, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-31 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural consequences of BMPR2 kinase domain mutations causing pulmonary arterial hypertension.
Sci Rep, 9, 2019
|
|
3JPV
 
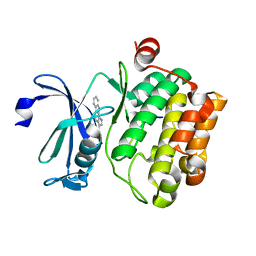 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a pyrrolo[2,3-a]carbazole ligand | | Descriptor: | 1,10-dihydropyrrolo[2,3-a]carbazole-3-carbaldehyde, Peptide (PIMTIDE) ARKRRRHPSGPPTA, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Akue-Gedu, R, Rossignol, E, Azzaro, S, Bain, J, Cohen, P, Prudhomme, M, Moreau, P, Amizon, F, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis, kinase inhibitory potencies, and in vitro antiproliferative evaluation of new pim kinase inhibitors.
J.Med.Chem., 52, 2009
|
|
3K2L
 
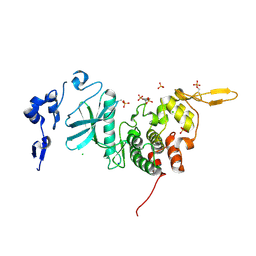 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) | | Descriptor: | CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2, SODIUM ION, ... | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Soundararajan, M, Krojer, T, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of Down Syndrome Kinases, DYRKs, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
3GG3
 
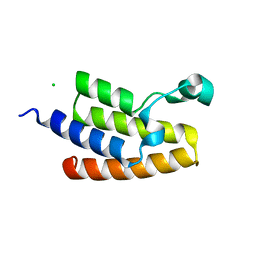 | | Crystal Structure of the Bromodomain of Human PCAF | | Descriptor: | CHLORIDE ION, Histone acetyltransferase PCAF | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, Rehana, K, Fedorov, O, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3P1D
 
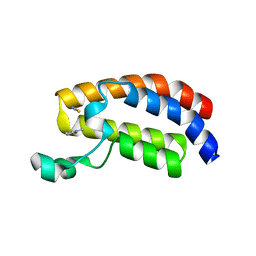 | | Crystal structure of the bromodomain of human CREBBP in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | 1-methylpyrrolidin-2-one, CREB-binding protein, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3P1C
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with acetylated lysine | | Descriptor: | CREB-binding protein, N(6)-ACETYLLYSINE, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
8SQJ
 
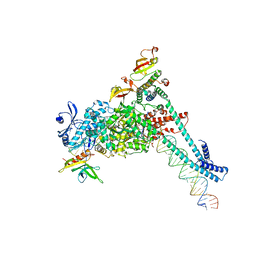 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQ9
 
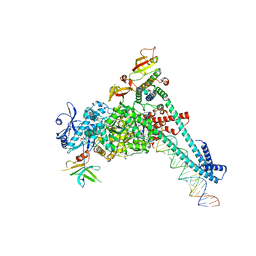 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQK
 
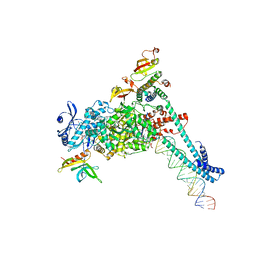 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
3O8D
 
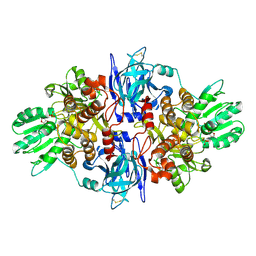 | |
3O8C
 
 | |
3O8B
 
 | |
3O8R
 
 | |
