8JKD
 
 | |
6BZ3
 
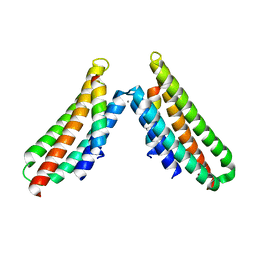 | | Complex structure of FAK FAT domain and DCC P3 motif | | Descriptor: | CALCIUM ION, Focal adhesion kinase 1, Netrin receptor DCC | | Authors: | Xu, S, Wang, J.-H. | | Deposit date: | 2017-12-22 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | The binding of DCC-P3 motif and FAK-FAT domain mediates the initial step of netrin-1/DCC signaling for axon attraction.
Cell Discov, 4, 2018
|
|
4E0R
 
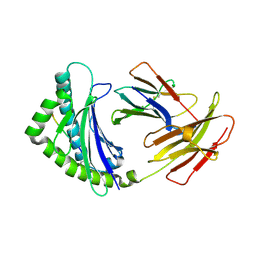 | | Structure of the chicken MHC class I molecule BF2*0401 | | Descriptor: | 8-MERIC PEPTIDE (FUS/TLS), Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
6O5I
 
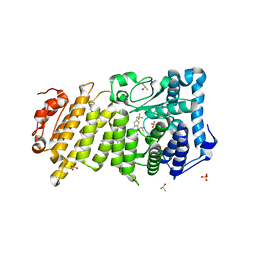 | | Menin in complex with MI-3454 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, SULFATE ION, ... | | Authors: | Linhares, B.M, Klossowski, S, Cierpicki, T, Grembecka, J. | | Deposit date: | 2019-03-03 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24025619 Å) | | Cite: | Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia.
J.Clin.Invest., 130, 2020
|
|
4LGI
 
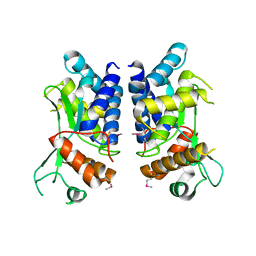 | | N-terminal truncated NleC structure | | Descriptor: | Uncharacterized protein | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5GMZ
 
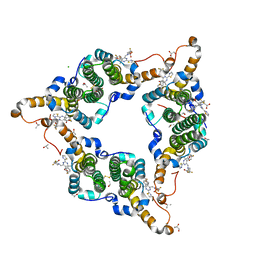 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
1GH2
 
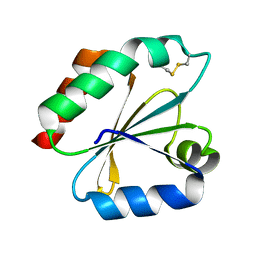 | | Crystal structure of the catalytic domain of a new human thioredoxin-like protein | | Descriptor: | THIOREDOXIN-LIKE PROTEIN | | Authors: | Jin, J, Chen, X, Guo, Q, Yuan, J, Qiang, B, Rao, Z. | | Deposit date: | 2000-11-01 | | Release date: | 2001-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the catalytic domain of a human thioredoxin-like protein.
Eur.J.Biochem., 269, 2002
|
|
4LGJ
 
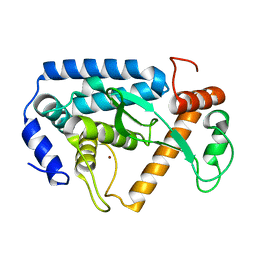 | | Crystal structure and mechanism of a type III secretion protease | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SIE
 
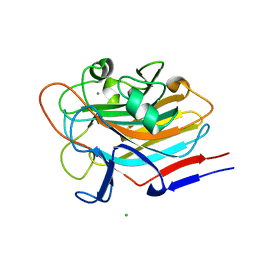 | | Structure of AAP A-domain (residues 351-605) from Staphylococcus epidermidis | | Descriptor: | Accumulation associated protein, CALCIUM ION, CHLORIDE ION | | Authors: | Atkin, K.E, Brentnall, A.S, Dodson, E.J, Whelan, F, Clark, L, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
4EJR
 
 | | Crystal structure of major capsid protein S domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Xu, F, Ma, J, Zhang, K, Wang, X, Sun, F. | | Deposit date: | 2012-04-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
8JPA
 
 | |
7SJK
 
 | | Structure of PLS A-domain (residues 391-656) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Pls Plasmin sensitive surface protein | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-18 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
7S03
 
 | |
6WF4
 
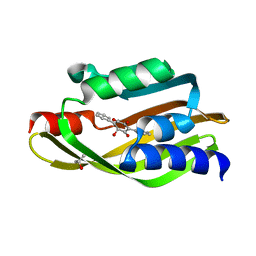 | | Crystal Structure of TerC Co-crystallized with Polyporic Acid | | Descriptor: | (2~5~S)-2~3~,2~5~,2~6~-trihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~(2~5~H)-one, ISOPROPYL ALCOHOL, Terfestatin Biosyntheis Enzyme C | | Authors: | Clinger, J.A, Miller, M.D, Hall, R.E, Zhang, Y, Elshahawi, S.I, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterization of two cooperative enzymes responsible for the stability
of p-terphenyls.
To be published
|
|
6KUU
 
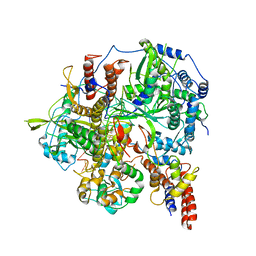 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
8WWP
 
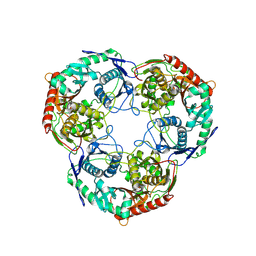 | | PNPase mutant of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
6PPO
 
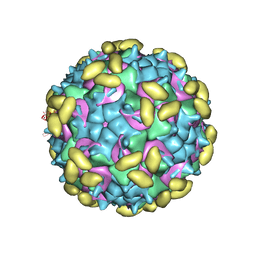 | | Rhinovirus C15 complexed with domain I of receptor CDHR3 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3, Capsid protein VP1, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1NFG
 
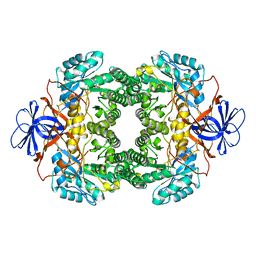 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
8WMH
 
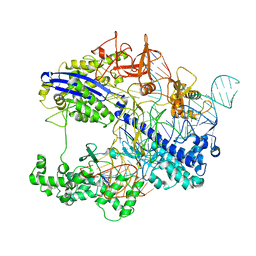 | | Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMM
 
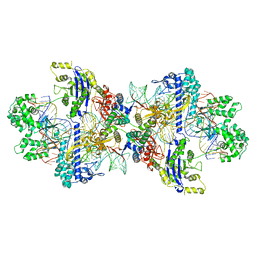 | | Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary) | | Descriptor: | MAGNESIUM ION, NTS, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMN
 
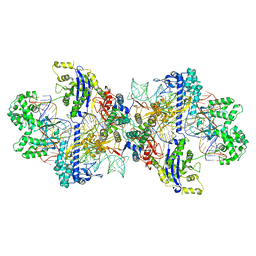 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary) | | Descriptor: | DNA (62-MER), MAGNESIUM ION, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WR4
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WXF
 
 | | PNPase of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-29 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
8X5D
 
 | |
6PSF
 
 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | Descriptor: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
