4F8P
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose | | Descriptor: | ACETATE ION, TERT-BUTYL FORMATE, beta-D-galactopyranose, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F8O
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with lactose and AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F8N
 
 | | X-ray structure of PsaA from Yersinia pestis, in complex with galactose and phosphate choline | | Descriptor: | CHLORIDE ION, GUANIDINE, PHOSPHOCHOLINE, ... | | Authors: | Bao, R, Esser, L, Xia, D. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2HB0
 
 | | Crystal Structure of CfaE, the Adhesive Subunit of CFA/I Fimbria of Enterotoxigenic Escherichia coli | | Descriptor: | CFA/I fimbrial subunit E, DI(HYDROXYETHYL)ETHER, MALONATE ION | | Authors: | Li, Y.F, Xia, D, Poole, S, Rasulova, F, Savarino, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
1FEP
 
 | | FERRIC ENTEROBACTIN RECEPTOR | | Descriptor: | FERRIC ENTEROBACTIN RECEPTOR | | Authors: | Buchanan, S.K, Smith, B.S, Ventatramani, L, Xia, D, Esser, L, Palnitkar, M, Chakraborty, R, Van Der Helm, D, Deisenhofer, J. | | Deposit date: | 1998-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the outer membrane active transporter FepA from Escherichia coli.
Nat.Struct.Biol., 6, 1999
|
|
8TI0
 
 | | ATP-1 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-07-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TBY
 
 | | Apo Bcs1, unsymmetrized | | Descriptor: | Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TPL
 
 | | ATP-2 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TP1
 
 | | ATP-2 state of Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
2FYN
 
 | | Crystal Structure Analysis of the double mutant Rhodobacter Sphaeroides bc1 complex | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, Cytochrome b, Cytochrome c1, ... | | Authors: | Esser, L, Xia, D. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Surface-modulated motion switch: Capture and release of iron-sulfur protein in the cytochrome bc1 complex.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7U9J
 
 | | Crystal structure of Mesothelin-207 fragment | | Descriptor: | GLYCEROL, Isoform 3 of Mesothelin, SULFATE ION | | Authors: | Zhan, J, Esser, L, Lin, D, Tang, W.K, Xia, D. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
7UED
 
 | | Crystal structure of full length mesothelin bound with MORAb-009 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Isoform 4 of Mesothelin, ... | | Authors: | Zhan, J, Esser, L, Lin, D, Tang, W.K, Xia, D. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
7U8C
 
 | | Crystal structure of Mesothelin C-terminal peptide-MORAb 15B6 FAB complex | | Descriptor: | MORab 15B6 Fab heavy chain, MORab 15B6 Fab light chain, Mesothelin, ... | | Authors: | Zhan, J, Esser, L, Xia, D. | | Deposit date: | 2022-03-08 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Highly active CAR T cells that bind to a juxtamembrane region of mesothelin and are not blocked by shed mesothelin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2QJK
 
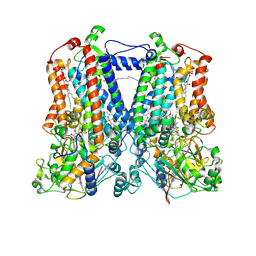 | | Crystal Structure Analysis of mutant rhodobacter sphaeroides bc1 with stigmatellin and antimycin | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Esser, L. | | Deposit date: | 2007-07-07 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides.
J.Biol.Chem., 283, 2008
|
|
7NAB
 
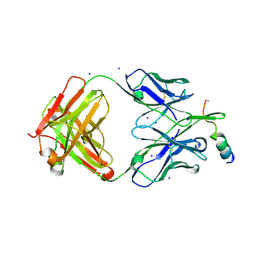 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
7MFU
 
 | | Crystal structure of synthetic nanobody (Sb14+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spike protein S1, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7MFV
 
 | | Crystal structure of synthetic nanobody (Sb16) | | Descriptor: | 1,2-ETHANEDIOL, Synthetic Nanobody #16 (Sb16) | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0H
 
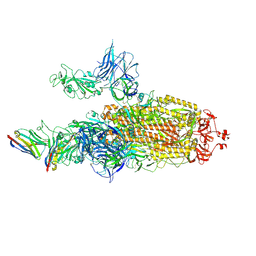 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0G
 
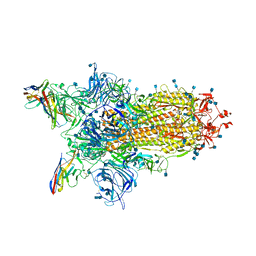 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7X1R
 
 | | Cryo-EM structure of human thioredoxin reductase bound by Au | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, Thioredoxin reductase 1, ... | | Authors: | He, Z.S, Cao, P, Cao, S.H, He, B, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Au4 cluster inhibits human thioredoxin reductase activity via specifically binding of Au to Cys189
Nano Today, 47, 2022
|
|
7KGK
 
 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb16, Sybody-16, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KLW
 
 | | Crystal structure of synthetic nanobody (Sb45+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | SB45, Synthetic Nanobody, SB68, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KGJ
 
 | | Crystal structure of synthetic nanobody (Sb45) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb45, Sybody-45, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
1AL0
 
 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | CAPSID PROTEIN GPF, SCAFFOLDING PROTEIN GPB, SCAFFOLDING PROTEIN GPD, ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1997-06-06 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a viral procapsid with molecular scaffolding.
Nature, 389, 1997
|
|
1CD3
 
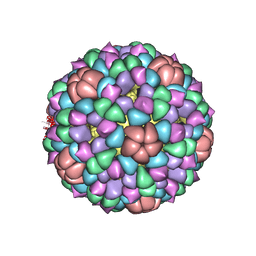 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | PROTEIN (CAPSID PROTEIN GPF), PROTEIN (SCAFFOLDING PROTEIN GPB), PROTEIN (SCAFFOLDING PROTEIN GPD), ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus phiX174.
J.Mol.Biol., 288, 1999
|
|
