3EZO
 
 | |
3IKF
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 717, imidazo[2,,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
6MB0
 
 | |
6MB1
 
 | | Crystal structure of N-myristoyl transferase (NMT) from Plasmodium vivax in complex with inhibitor IMP-1002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-{4-fluoro-2-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]phenyl}-1-methyl-1H-indazol-3-yl)-N,N-dimethylmethanamine, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Identification of Resistance Breaking Antimalarial N‐Myristoyltransferase Inhibitors.
Cell Chem Biol, 26, 2019
|
|
6MAZ
 
 | |
6MAY
 
 | |
6N1C
 
 | |
4E51
 
 | |
3K2X
 
 | |
3K14
 
 | | Co-crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 535, ethyl 3-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole-2-carboxylate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
3K9W
 
 | |
3MD0
 
 | |
3MBM
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with cytosine and FoL fragment 717, imidazo[2,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
4NHF
 
 | |
4P9A
 
 | | X-ray Crystal Structure of PA protein from Influenza strain H7N9 | | Descriptor: | GLYCEROL, Polymerase PA | | Authors: | Fairman, J.W, Moen, S.O, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1.
Sci Rep, 4, 2014
|
|
4O2D
 
 | |
4O3V
 
 | |
4Q6U
 
 | |
4RGB
 
 | |
6AUB
 
 | |
4EFI
 
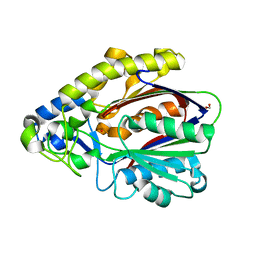 | | Crystal Structure of 3-oxoacyl-(Acyl-carrier protein) Synthase from Burkholderia Xenovorans LB400 | | Descriptor: | 3-oxoacyl-(Acyl-carrier protein) synthase, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Craig, T.K, Abendroth, J, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
4H4G
 
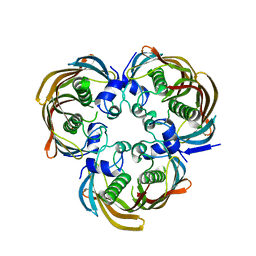 | | Crystal Structure of (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase from Burkholderia thailandensis E264 | | Descriptor: | (3R)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase | | Authors: | Craig, T.K, Edwards, T.E, Staker, B, Stewart, L, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-09-17 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
6AUA
 
 | |
4F2G
 
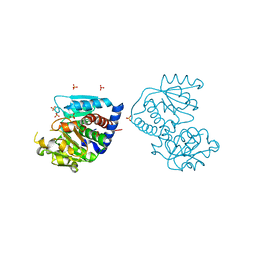 | | The Crystal Structure of Ornithine carbamoyltransferase from Burkholderia thailandensis E264 | | Descriptor: | 1,2-ETHANEDIOL, Ornithine carbamoyltransferase 1, PHOSPHATE ION | | Authors: | Craig, T.K, Fox, D, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
3F0D
 
 | |
