4KQL
 
 | | Hin GlmU bound to WG578 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-(4-{[3-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-5-methoxybenzoyl]amino}phenyl)pyridine-2-carboxamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-15 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
4KW5
 
 | | M. tuberculosis DprE1 in complex with inhibitor TCA1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Identification of a small molecule with activity against drug-resistant and persistent tuberculosis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3PN1
 
 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | Authors: | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
3OHT
 
 | | Crystal Structure of Salmo Salar p38alpha | | Descriptor: | N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, SULFATE ION, p38a | | Authors: | Rothweiler, U, Johnson, K, Engh, R.A. | | Deposit date: | 2010-08-18 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | p38alpha MAP kinase dimers with swapped activation segments and a novel catalytic loop conformation
J.Mol.Biol., 411, 2011
|
|
8FUR
 
 | | Crystal structure of human IDO1 with compound 11 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-methylphenyl)-N'-[(1P,2'P)-4-propoxy-5-propyl-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-yl]urea | | Authors: | Critton, D.A, Lewis, H.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and biological evaluation of biaryl alkyl ethers as inhibitors of IDO1.
Bioorg.Med.Chem.Lett., 88, 2023
|
|
6TCZ
 
 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 | | Descriptor: | Proteasome endopeptidase complex, Proteasome subunit alpha type, Proteasome subunit beta, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
6TD5
 
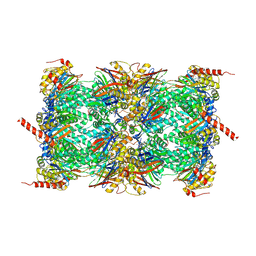 | | Leishmania tarentolae proteasome 20S subunit complexed with LXE408 and bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Srinivas, H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and Characterization of Clinical Candidate LXE408 as a Kinetoplastid-Selective Proteasome Inhibitor for the Treatment of Leishmaniases.
J.Med.Chem., 63, 2020
|
|
6X2J
 
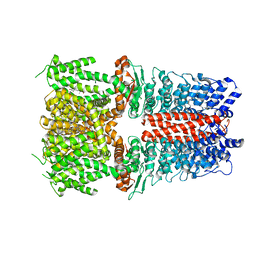 | | Structure of human TRPA1 in complex with agonist GNE551 | | Descriptor: | 5-amino-1-[(4-bromo-2-fluorophenyl)methyl]-N-(2,5-dimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Chen, H. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A Non-covalent Ligand Reveals Biased Agonism of the TRPA1 Ion Channel.
Neuron, 109, 2021
|
|
1GJY
 
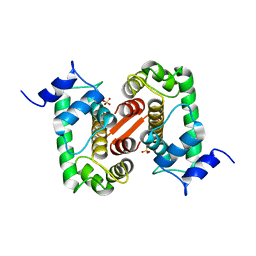 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
3IJJ
 
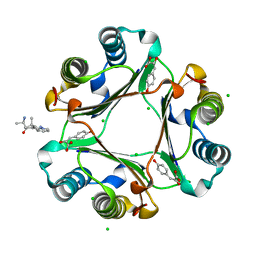 | | Ternary Complex of Macrophage Migration Inhibitory Factor (MIF) Bound Both to 4-hydroxyphenylpyruvate and to the Allosteric Inhibitor AV1013 (R-stereoisomer) | | Descriptor: | (2E)-2-hydroxy-3-(4-hydroxyphenyl)prop-2-enoic acid, (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ... | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IJG
 
 | | Macrophage Migration Inhibitory Factor (MIF) Bound to the (R)-Stereoisomer of AV1013 | | Descriptor: | (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5FX5
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | GLYCEROL, RHINOVIRUS 3C PROTEASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5FED
 
 | | EGFR kinase domain in complex with a covalent aminobenzimidazole inhibitor. | | Descriptor: | Epidermal growth factor receptor, ~{N}-[7-methyl-1-[(3~{R})-1-propanoylazepan-3-yl]benzimidazol-2-yl]-3-(trifluoromethyl)benzamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
5FEE
 
 | | EGFR kinase domain T790M mutant in complex with a covalent aminobenzimidazole inhibitor. | | Descriptor: | Epidermal growth factor receptor, ~{N}-[7-methyl-1-[(3~{R})-1-propanoylazepan-3-yl]benzimidazol-2-yl]-3-(trifluoromethyl)benzamide | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (R,E)-N-(7-Chloro-1-(1-[4-(dimethylamino)but-2-enoyl]azepan-3-yl)-1H-benzo[d]imidazol-2-yl)-2-methylisonicotinamide (EGF816), a Novel, Potent, and WT Sparing Covalent Inhibitor of Oncogenic (L858R, ex19del) and Resistant (T790M) EGFR Mutants for the Treatment of EGFR Mutant Non-Small-Cell Lung Cancers.
J.Med.Chem., 59, 2016
|
|
5IEN
 
 | | Structure of CDL2.2, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.2, GLYCEROL | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5FX6
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | RHINOVIRUS 3C PROTEASE, ethyl (4R)-4-[[(2S,4S)-1-[(2S)-3-methyl-2-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]butanoyl]-4-phenyl-pyrrolidin-2-yl]carbonylamino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5I5K
 
 | |
5IEO
 
 | | Structure of CDL2.3a, a computationally designed Vitamin-D3 binder | | Descriptor: | 1,2-ETHANEDIOL, 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3a | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IER
 
 | |
5IEP
 
 | | Structure of CDL2.3b, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3b | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IF6
 
 | |
7JUP
 
 | | Structure of human TRPA1 in complex with antagonist compound 21 | | Descriptor: | 1-({3-[(3R,5R)-5-(4-fluorophenyl)oxolan-3-yl]-1,2,4-oxadiazol-5-yl}methyl)-7-methyl-1,7-dihydro-6H-purin-6-one, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L. | | Deposit date: | 2020-08-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Tetrahydrofuran-Based Transient Receptor Potential Ankyrin 1 (TRPA1) Antagonists: Ligand-Based Discovery, Activity in a Rodent Asthma Model, and Mechanism-of-Action via Cryogenic Electron Microscopy.
J.Med.Chem., 64, 2021
|
|
5WO4
 
 | | JAK1 complexed with compound 28 | | Descriptor: | 3-[(4-chloro-3-methoxyphenyl)amino]-1-[(3R,4S)-4-cyanooxan-3-yl]-1H-pyrazole-4-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lesburg, C.A, Patel, S.B. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Discovery of 3-((4-Chloro-3-methoxyphenyl)amino)-1-((3R,4S)-4-cyanotetrahydro-2H-pyran-3-yl)-1H-pyrazole-4-carboxamide, a Highly Ligand Efficient and Efficacious Janus Kinase 1 Selective Inhibitor with Favorable Pharmacokinetic Properties.
J. Med. Chem., 60, 2017
|
|
4J9A
 
 | | Engineered Digoxigenin binder DIG10.3 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.3 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-15 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
4J8T
 
 | | Engineered Digoxigenin binder DIG10.2 | | Descriptor: | DIGOXIGENIN, Engineered Digoxigenin binder protein DIG10.2 | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of ligand-binding proteins with high affinity and selectivity.
Nature, 501, 2013
|
|
