8I0Y
 
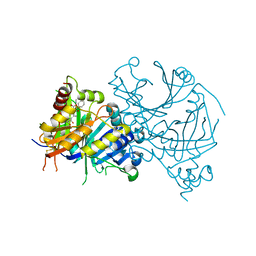 | | Crystal structure of AtHPPD-Y191713 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, ethyl 5-methyl-6-[(2-methyl-3-oxidanylidene-1H-pyrazol-4-yl)carbonyl]-4-oxidanylidene-3-phenyl-quinazoline-2-carboxylate | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of AtHPPD-Y191713 complex
To Be Published
|
|
8I2S
 
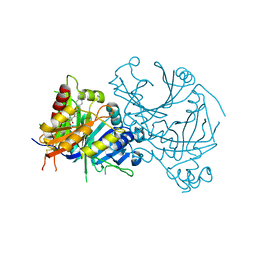 | | Crystal structure of AtHPPD-Y18979 complex | | Descriptor: | 1,5-dimethyl-3-(naphthalen-2-ylmethyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2023-01-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Crystal structure of AtHPPD-Y18979 complex
To Be Published
|
|
3K3Q
 
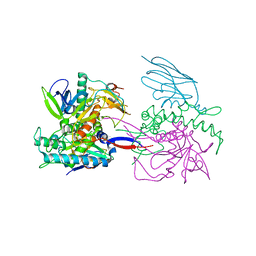 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | Descriptor: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | Authors: | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | Deposit date: | 2009-10-04 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
5WEQ
 
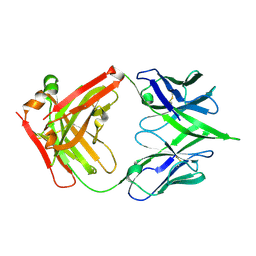 | | The crystal structure of a MR78 mutant | | Descriptor: | MR78 mutant Fab heavy chain, MR78 mutant light chain | | Authors: | Dong, J, Williamson, L.E, Crowe, J.E. | | Deposit date: | 2017-07-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Non-local Interactions between CDR Loops in Binding Affinity of MR78 Antibody to Marburg Virus Glycoprotein.
Structure, 25, 2017
|
|
4RVZ
 
 | | Crystal structure of tRNA fluorescent labeling enzyme | | Descriptor: | MAGNESIUM ION, N-(4-aminobutyl)-2-azidoacetamide, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Dong, J, Li, F, Wang, J, Gong, W. | | Deposit date: | 2014-11-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A covalent approach for site-specific RNA labeling in Mammalian cells.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TQN
 
 | |
5JRP
 
 | | crystal structure of monoclonal antibody MR78 Fab | | Descriptor: | SODIUM ION, marberg virus monoclonal antibody MR78 Fab heavy chain, marberg virus monoclonal antibody MR78 Fab light chain | | Authors: | Dong, J, Crowe, J. | | Deposit date: | 2016-05-06 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Non-local Interactions between CDR Loops in Binding Affinity of MR78 Antibody to Marburg Virus Glycoprotein.
Structure, 25, 2017
|
|
6P9J
 
 | | crystal structure of human anti staphylococcus aureus antibody STAU-229 Fab | | Descriptor: | TRIS(HYDROXYETHYL)AMINOMETHANE, human anti staphylococcus aureus antibody STAU-229 Fab heavy chain, human anti staphylococcus aureus antibody STAU-229 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
6P9I
 
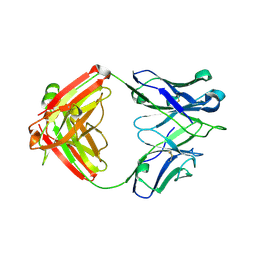 | | crystal structure of human anti staphylococcus aureus antibody STAU-399 Fab | | Descriptor: | human anti staphylococcus aureus antibody STAU-399 Fab heavy chain, human anti staphylococcus aureus antibody STAU-399 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
6P9H
 
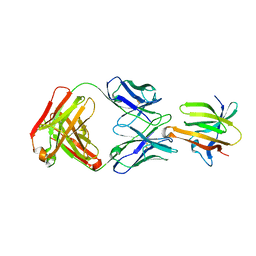 | |
3K8A
 
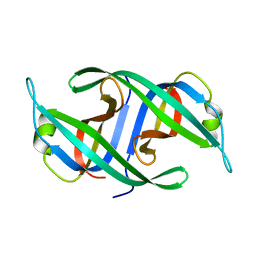 | | Neisseria gonorrhoeae PriB | | Descriptor: | Putative primosomal replication protein | | Authors: | Lopper, M.E, Dong, J, George, N.P, Duckett, K.L, DeBeer, M.A. | | Deposit date: | 2009-10-14 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Neisseria gonorrhoeae PriB reveals mechanistic differences among bacterial DNA replication restart pathways
Nucleic Acids Res., 38, 2010
|
|
5H85
 
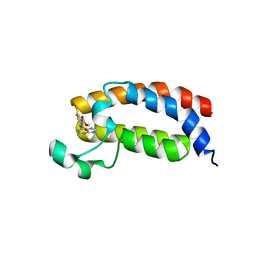 | |
6DLB
 
 | |
6DLA
 
 | |
6DL8
 
 | |
5HCL
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with DMA | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N},~{N}-dimethylethanamide | | Authors: | Dong, J, Weber, F.E, Caflisch, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N,N Dimethylacetamide a drug excipient that acts as bromodomain ligand for osteoporosis treatment.
Sci Rep, 7, 2017
|
|
7XVH
 
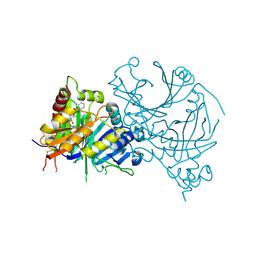 | | Crystal structure of AtHPPD-Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
8X6Q
 
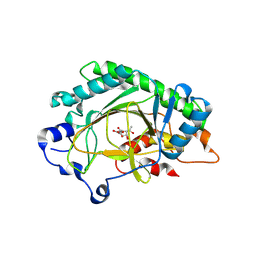 | | Crystal structure of OsHSL1 L204F/F298L/I335F complexed with 2-acetyl-cyclohexane-2,4-dione | | Descriptor: | 2-OXOGLUTARIC ACID, 2-ethanoyl-3-oxidanyl-cyclohex-2-en-1-one, COBALT (II) ION, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-11-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7D
 
 | |
8X74
 
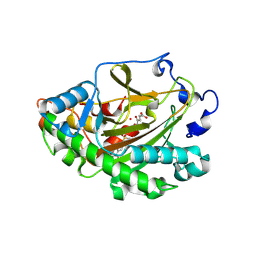 | | Crystal structure of ZmHSL1A complexed with mesotrione | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[(4-methylsulfonyl-2-nitro-phenyl)-oxidanyl-methylidene]cyclohexane-1,3-dione, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XC3
 
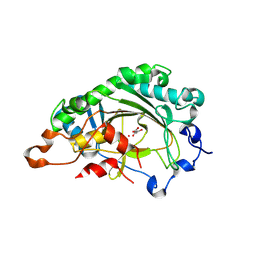 | | Crystal structure of ZmHSL1A-MBQ complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 2-OXOGLUTARIC ACID, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, ... | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7C
 
 | | Crystal structure of ZmHSL1A | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-11-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8HOW
 
 | | Crystal structure of AtHPPD-Y191052 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(2-phenylethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-12-11 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
4GF6
 
 | | crystal structure of GFP with cuprum bound at the Incorporated metal Chelating Amino Acid PYZ151 | | Descriptor: | CALCIUM ION, COPPER (II) ION, green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4GES
 
 | | crystal structure of GFP-TYR151PYZ with an unnatural amino acid incorporation | | Descriptor: | Green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
