5VS6
 
 | | Structure of DUB complex | | Descriptor: | ACETATE ION, GLYCEROL, N-[3-({4-hydroxy-1-[(3R)-3-phenylbutanoyl]piperidin-4-yl}methyl)-4-oxo-3,4-dihydroquinazolin-7-yl]-3-(4-methylpiperazin-1-yl)propanamide, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
5VSK
 
 | | Structure of DUB complex | | Descriptor: | 7-chloro-3-({4-hydroxy-1-[(3S)-3-phenylbutanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2017-05-11 | | Release date: | 2017-12-20 | | Last modified: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7.
Cell Chem Biol, 24, 2017
|
|
7ELJ
 
 | |
4ZH2
 
 | |
4ZH3
 
 | |
5ULT
 
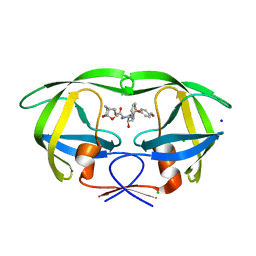 | | HIV-1 wild Type protease with GRL-100-13A (a Crown-like Oxotricyclic Core as the P2-Ligand with the sulfonamide isostere as the P2' group) | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and Development of Highly Potent HIV-1 Protease Inhibitors with a Crown-Like Oxotricyclic Core as the P2-Ligand To Combat Multidrug-Resistant HIV Variants.
J. Med. Chem., 60, 2017
|
|
3HSA
 
 | |
7UE1
 
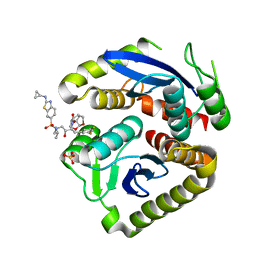 | | HIV-1 Integrase Catalytic Core Domain Mutant (KGD) in Complex with Inhibitor GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Integrase, SULFATE ION | | Authors: | Aoki, M, Aoki-Ogata, H, Bulut, H, Hayashi, H, Davis, D, Hasegawa, K, Yarchoan, R, Ghosh, A.K, Pau, A.K, Mitsuya, H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants.
Sci Adv, 9, 2023
|
|
5CXT
 
 | |
3KK7
 
 | |
4ZCH
 
 | | Single-chain human APRIL-BAFF-BAFF Heterotrimer | | Descriptor: | TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, Tumor necrosis factor ligand superfamily member 13,Tumor necrosis factor ligand superfamily member 13B,Tumor necrosis factor ligand superfamily member 13B | | Authors: | Lammens, A, Jiang, X, Maskos, K, Schneider, P. | | Deposit date: | 2015-04-16 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Stoichiometry of Heteromeric BAFF and APRIL Cytokines Dictates Their Receptor Binding and Signaling Properties.
J.Biol.Chem., 290, 2015
|
|
6JR3
 
 | |
6MCR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
6MCS
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
3NL9
 
 | |
2OOK
 
 | |
2PV7
 
 | |
2Q3L
 
 | |
2QTP
 
 | |
2RA9
 
 | |
5V4Y
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-09510 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Delino, N.S, Das, D, Kaufman, J.D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2017-03-11 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GRL-09510, a Unique P2-Crown-Tetrahydrofuranylurethane -Containing HIV-1 Protease Inhibitor, Maintains Its Favorable Antiviral Activity against Highly-Drug-Resistant HIV-1 Variants in vitro.
Sci Rep, 7, 2017
|
|
4L2W
 
 | |
2RE3
 
 | |
3BYQ
 
 | |
3CM1
 
 | |
