5C1V
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CATALYTIC SUBUNIT OF HUMAN CALCINEURIN | | Descriptor: | FE (III) ION, PHOSPHATE ION, Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform, ... | | Authors: | Guasch, A, Fita, I, Perez-Luque, R, Aparicio, D, Aranguren-Ibanez, A, Perez-Riba, M. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcineurin Undergoes a Conformational Switch Evoked via Peptidyl-Prolyl Isomerization.
Plos One, 10, 2015
|
|
5C9F
 
 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, CHLORIDE ION, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3QGM
 
 | | p-nitrophenyl phosphatase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Osipiuk, J, Zheng, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-01-24 | | Release date: | 2011-02-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
3HDJ
 
 | | The crystal structure of probable ornithine cyclodeaminase from Bordetella pertussis Tohama I | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Tan, K, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of probable ornithine cyclodeaminase from Bordetella pertussis Tohama I
To be Published
|
|
2FVJ
 
 | | A novel anti-adipogenic partial agonist of peroxisome proliferator-activated receptor-gamma (PPARG) recruits pparg-coactivator-1 alpha (PGC1A) but potentiates insulin signaling in vitro | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXY-4-{[4-(2-METHOXYPHENYL)PIPERIDIN-1-YL]METHYL}ISOQUINOLINE, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Benz, J, Burgermeister, E, Flament, A, Schnoebelen, A, Stihle, M, Gsell, B, Rufer, A, Ruf, A, Kuhn, B, Maerki, H.P, Mizrahi, J, Sebokova, E, Niesor, E, Meyer, M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A novel partial agonist of peroxisome proliferator-activated receptor-gamma (PPARgamma) recruits PPARgamma-coactivator-1alpha, prevents triglyceride accumulation, and potentiates insulin signaling in vitro
Mol.Endocrinol., 20, 2006
|
|
4J9B
 
 | |
5CN9
 
 | | Ultrafast dynamics in myoglobin: 0.4 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
3FHM
 
 | | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Zhang, R, Cui, H, Kudritsdka, M, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens
To be Published
|
|
5CNG
 
 | | ultrafast dynamics in myoglobin: 150 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
3HKO
 
 | | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40 | | Descriptor: | Calcium/calmodulin-dependent protein kinase with a kinase domain and 2 calmodulin-like EF hands, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Wasney, G, Vedadi, M, MacKenzie, F, Kozieradzki, I, Cossar, D, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Botchkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-25 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a cdpk kinase domain from cryptosporidium Parvum, cgd7_40
To be Published
|
|
2DXQ
 
 | | Putative acetyltransferase from Agrobacterium tumefaciens str. C58 | | Descriptor: | Acetyltransferase, PHOSPHATE ION | | Authors: | Binkowski, T.A, Xu, X, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Putative acetyltransferase from Agrobacterium tumefaciens str. C58
To be published
|
|
2PAS
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Padilla, A, Cave, A, Parello, J, Etienne, G, Baldellon, C. | | Deposit date: | 1994-03-22 | | Release date: | 1994-06-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin
To be Published
|
|
1XGK
 
 | | CRYSTAL STRUCTURE OF N12G AND A18G MUTANT NMRA | | Descriptor: | CHLORIDE ION, GLYCEROL, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, ... | | Authors: | Lamb, H.K, Ren, J, Park, A, Johnson, C, Leslie, K, Cocklin, S, Thompson, P, Mee, C, Cooper, A, Stammers, D.K, Hawkins, A.R. | | Deposit date: | 2004-09-17 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis
Protein Sci., 13, 2004
|
|
3JCZ
 
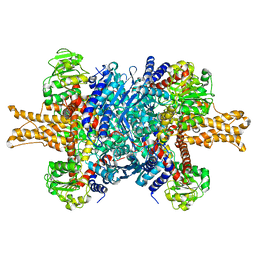 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
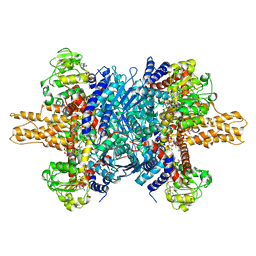 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JQ4
 
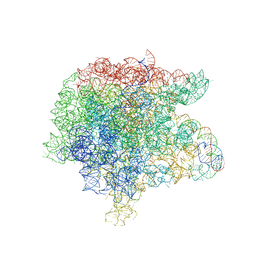 | | The structure of the complex of the large ribosomal subunit from D. Radiodurans with the antibiotic lankacidin | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, N-[(1S,2R,3E,5E,7S,9E,11E,13S,15R,19R)-7,13-dihydroxy-1,4,10,19-tetramethyl-17,18-dioxo-16-oxabicyclo[13.2.2]nonadeca-3,5,9,11-tetraen-2-yl]-2-oxopropanamide | | Authors: | Auerbach-Nevo, T, Mermershtain, I, Davidovich, C, Bashan, A, Rozenberg, H, Yonath, A. | | Deposit date: | 2009-09-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5G15
 
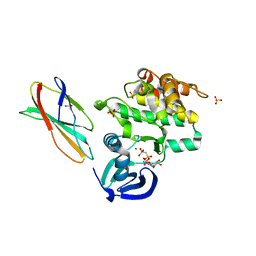 | | Structure Aurora A (122-403) bound to activating monobody Mb1 and AMPPCP | | Descriptor: | AURORA A KINASE, MAGNESIUM ION, MB1 MONOBODY, ... | | Authors: | Zorba, A, Kutter, S, Kern, D, Koide, S, Koide, A. | | Deposit date: | 2016-03-23 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2YLY
 
 | | Sulfonamides as selective Estrogen Receptor beta Agonists. | | Descriptor: | ESTROGEN RECEPTOR BETA, N-cyclopropyl-4-oxidanyl-N-[(2R)-2-oxidanyl-2-phenyl-propyl]benzenesulfonamide, SULFATE ION | | Authors: | Roberts, L.R, Armour, D, Barker, C, Bazin, R, Bess, K, Brown, A, Favor, D, Ellis, D, MacKenny, M, Pullen, N, Stennett, A, Strand, L, Styles, M, Phillips, C. | | Deposit date: | 2011-06-06 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sulfonamides as Selective Oestrogen Receptor Beta Agonists.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5FZI
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with MC3095 | | Descriptor: | 1,2-ETHANEDIOL, 6-oxo-2-[(2-oxo-2-phenylethyl)sulfanyl]-1,6-dihydropyrimidine-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Rotili, D, Mai, A, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Mc3095
To be Published
|
|
3K2J
 
 | | Crystal Structure of the 3rd Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | CHLORIDE ION, Protein polybromo-1, SULFATE ION | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Chaikuad, A, Pike, A.C.W, Krojer, T, Sethi, R, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the 3rd Bromodomain of Human Poly-bromodomain containing protein 1 (PB1)
To be Published
|
|
1XPP
 
 | | Crystal Structure of TA1416,DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum | | Descriptor: | ACETIC ACID, DNA-directed RNA polymerase subunit L, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of TA1416, DNA-directed RNA polymerase subunit L, from Thermoplasma acidophilum
To be Published
|
|
3QTD
 
 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
3J6B
 
 | | Structure of the yeast mitochondrial large ribosomal subunit | | Descriptor: | 21S ribosomal RNA, 54S ribosomal protein IMG1, mitochondrial, ... | | Authors: | Amunts, A, Brown, A, Bai, X.C, Llacer, J.L, Hussain, T, Emsley, P, Long, F, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the yeast mitochondrial large ribosomal subunit.
Science, 343, 2014
|
|
5FEX
 
 | | HydE from T. maritima in complex with Se-adenosyl-L-selenocysteine (tfinal of the reaction) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
1XMK
 
 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
