3LYA
 
 | |
3LYL
 
 | | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-27 | | Release date: | 2010-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis
TO BE PUBLISHED
|
|
4W97
 
 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4USM
 
 | | WcbL complex with glycerol bound to sugar site | | Descriptor: | CHLORIDE ION, GLYCEROL, PUTATIVE SUGAR KINASE | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
6JDW
 
 | |
4UT4
 
 | | Burkholderia pseudomallei heptokinase WcbL, D-mannose complex. | | Descriptor: | CHLORIDE ION, PUTATIVE SUGAR KINASE, alpha-D-mannopyranose | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
4UX8
 
 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
6Q0N
 
 | | Structure of the Erbin PDB domain in complex with a high-affinity peptide | | Descriptor: | Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6Q0S
 
 | |
6Q0M
 
 | | Structure of Erbin PDZ derivative E-14 with a high-affinity peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6Q0U
 
 | | Structure of the Erbin PDZ variant E-6a with a high-affinity C-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6Q39
 
 | | Complex of Arginase 2 with Example 49 | | Descriptor: | 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-1-[[(2~{S})-piperidin-2-yl]methyl]pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, mitochondrial, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
6QD6
 
 | | Molecular scaffolds expand the nanobody toolkit for cryo-EM applications: crystal structure of Mb-cHopQ-Nb207 | | Descriptor: | CHLORIDE ION, Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207,Outer membrane protein,Mb-cHopQ-Nb207 | | Authors: | Uchanski, T, Masiulis, S, Fischer, B, Kalichuk, V, Wohlkonig, A, Zogg, T, Remaut, H, Vranken, W, Aricescu, A.R, Pardon, E, Steyaert, J. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM
Nat.Methods, 18, 2021
|
|
6QAT
 
 | | Crystal structure of ULK2 in complexed with hesperadin | | Descriptor: | N-{(3Z)-2-oxo-3-[phenyl({4-[(piperidin-1-yl)methyl]phenyl}amino)methylidene]-2,3-dihydro-1H-indol-5-yl}ethanesulfonamide, Serine/threonine-protein kinase ULK2 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QIS
 
 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model II) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3'), SULFATE ION | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6D24
 
 | | Trypanosoma cruzi Glucose-6-P Dehydrogenase in complex with G6P | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Botti, H, Ortiz, C, Comini, M.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Glucose-6-Phosphate Dehydrogenase from the Human Pathogen Trypanosoma cruzi Evolved Unique Structural Features to Support Efficient Product Formation.
J.Mol.Biol., 431, 2019
|
|
6JHD
 
 | | Solution structure of IFN alpha8 | | Descriptor: | Interferon alpha-8 | | Authors: | Ken-ichi, A, Shigeyuki, M. | | Deposit date: | 2019-02-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structure of interferon alpha8:
Novel insights into the preferred interaction with IFNAR2 among its subtypes
To Be Published
|
|
6Q42
 
 | | Crystal Structure of Human Pancreatic Phospholipase A2 | | Descriptor: | CHLORIDE ION, Phospholipase A2 | | Authors: | Saul, F, Haouz, A, Lambeau, G, Theze, J. | | Deposit date: | 2018-12-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PLA2G1B is involved in CD4 anergy and CD4 lymphopenia in HIV-infected patients.
J.Clin.Invest., 130, 2020
|
|
6QGK
 
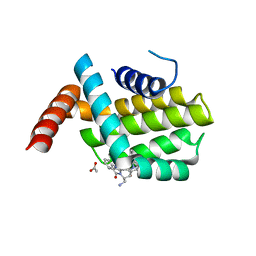 | | Structure of human Bcl-2 in complex with THIQ-phenyl pyrazole compound | | Descriptor: | 1-[2-[[(3~{S})-3-(aminomethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]phenyl]-~{N},~{N}-dibutyl-5-methyl-pyrazole-3-carboxamide, ACETATE ION, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QAS
 
 | | Crystal structure of ULK1 in complexed with PF-03814735 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QAU
 
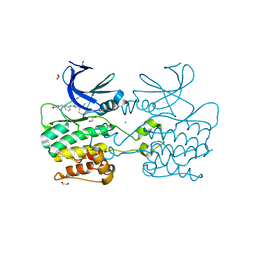 | | Crystal structure of ULK2 in complexed with MRT67307 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6D23
 
 | | GLUCOSE-6-P DEHYDROGENASE (APO FORM) FROM TRYPANOSOMA CRUZI | | Descriptor: | CHLORIDE ION, GLYCEROL, Glucose-6-phosphate 1-dehydrogenase, ... | | Authors: | Botti, H, Ortiz, C, Comini, M.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Glucose-6-Phosphate Dehydrogenase from the Human Pathogen Trypanosoma cruzi Evolved Unique Structural Features to Support Efficient Product Formation.
J.Mol.Biol., 431, 2019
|
|
6QDR
 
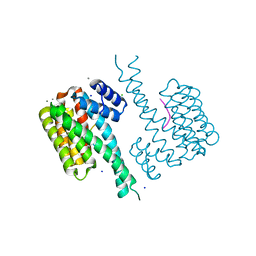 | | Crystal structure of 14-3-3sigma in complex with a PAK6 pT99 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Kaplan, A, Fournier, A.E, Ottman, C. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.615 Å) | | Cite: | Polypharmacological Perturbation of the 14-3-3 Adaptor Protein Interactome Stimulates Neurite Outgrowth.
Cell Chem Biol, 27, 2020
|
|
6QGD
 
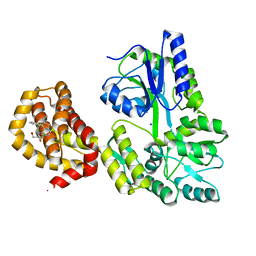 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGG
 
 | | Structure of human Bcl-2 in complex with analogue of ABT-737 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, [(3~{R})-3-[[4-[[4-[4-[[2-(4-chlorophenyl)phenyl]methyl]piperazin-1-yl]phenyl]carbonylsulfamoyl]-2-nitro-phenyl]amino]-4-phenylsulfanyl-butyl]-(2-hydroxy-2-oxoethyl)-dimethyl-azanium | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
