8ZHD
 
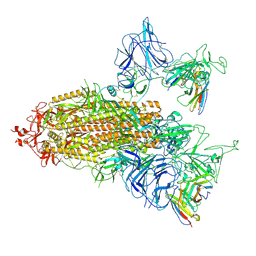 | | SARS-CoV-2 spike trimer (6P) in complex with two R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHI
 
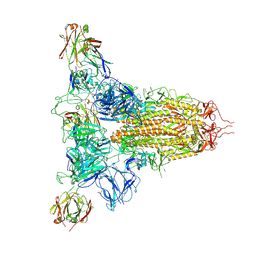 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.05 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHJ
 
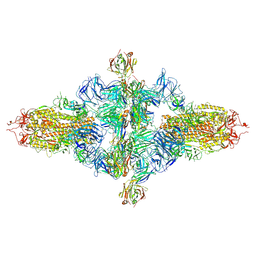 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C1 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
1KBR
 
 | |
9INR
 
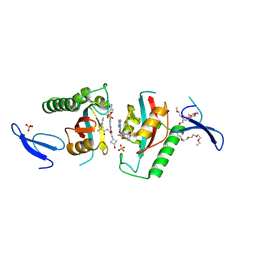 | | Crystal structure of PIN1 in complex with inhibitor C3 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Zhang, L.J, Zhang, L.Y. | | Deposit date: | 2024-07-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Re-Evaluating PIN1 as a Therapeutic Target in Oncology Using Neutral Inhibitors and PROTACs.
J.Med.Chem., 67, 2024
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
5XFF
 
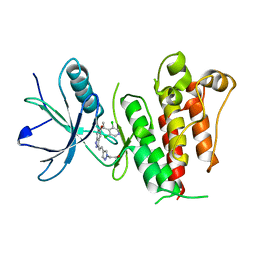 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550L) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
6QSU
 
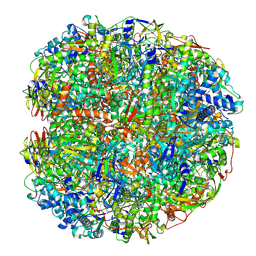 | | Helicobacter pylori urease with BME bound in the active site | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2019-02-22 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
8Y7L
 
 | |
6ZJA
 
 | | Helicobacter pylori urease with inhibitor bound in the active site | | Descriptor: | 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyacetamide, NICKEL (II) ION, Urease subunit alpha, ... | | Authors: | Luecke, H, Cunha, E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-12-23 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 angstrom resolution.
Nat Commun, 12, 2021
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GPY
 
 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
5C5S
 
 | |
6EFB
 
 | | GspB Siglec domain | | Descriptor: | CALCIUM ION, GLYCEROL, SK150 siglec + Unique, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EF7
 
 | | GspB Siglec domain | | Descriptor: | CALCIUM ION, Platelet binding protein GspB | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFA
 
 | | GspB Siglec + Unique domains | | Descriptor: | CALCIUM ION, Platelet binding protein GspB | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6ER4
 
 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,6 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
6EF9
 
 | | GspB Siglec domain | | Descriptor: | FORMIC ACID, Platelet binding protein GspB - Siglec domain, SODIUM ION | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFD
 
 | | Hsa Siglec and Unique domains in complex with the sialyl T antigen trisaccharide | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFF
 
 | | NCTC10712 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6ER2
 
 | |
6WFK
 
 | | Crystal structure of human Naa50 in complex with CoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFN
 
 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
8DEA
 
 | |
8DG6
 
 | |
