4MUB
 
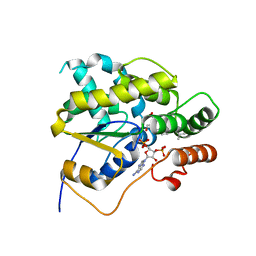 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase/Oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2S)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Valentim, C.L.L, Cioli, D, Chevalier, F.D, Cao, X, Taylor, A.B, Holloway, S.P, Pica-Mattoccia, L, Guidi, A, Basso, A, Tsai, I.J, Berriman, M, Carvalho-Queiroz, C, Almeida, M, Aguilar, H, Frantz, D.E, Hart, P.J, Anderson, T.J.C, LoVerde, P.T. | | Deposit date: | 2013-09-21 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and molecular basis of drug resistance and species-specific drug action in schistosome parasites.
Science, 342, 2013
|
|
6B3N
 
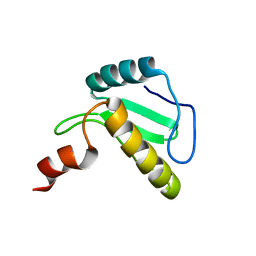 | | Solution structure of the N-terminal domain of the effector NleG5-1 from Escherichia coli O157:H7 str. Sakai | | Descriptor: | NleG5-1 | | Authors: | Valleau, D, Houliston, S, Lemak, A, Anderson, W.F, Arrowsmith, C, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of the effector NleG5-1 from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
6YXW
 
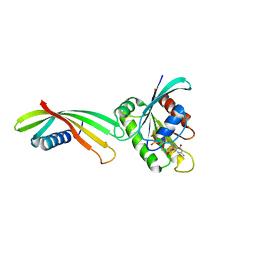 | | Affimer K3 - KRAS protein complex | | Descriptor: | Affimer K3, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
6FI0
 
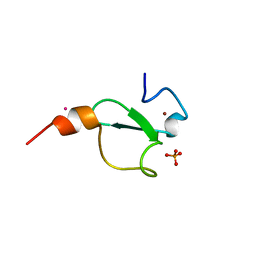 | | Crystal structure of BAZ2A PHD zinc finger in complex with Fr 19 | | Descriptor: | 2-azanyl-1-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6QA2
 
 | | R80A MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | Nucleoside diphosphate kinase, SULFATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Dautant, A, Henri, J, Wales, T.E, Meyer, P, Engen, J.R, Georgescauld, F. | | Deposit date: | 2018-12-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Remodeling of the Binding Site of Nucleoside Diphosphate Kinase Revealed by X-ray Structure and H/D Exchange.
Biochemistry, 58, 2019
|
|
6BU6
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor | | Descriptor: | 4,4'-(2-aminopyridine-3,5-diyl)bis(2,6-difluorophenol), CHLORIDE ION, GLYCEROL, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Santiago, A.S, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-08 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor
To Be Published
|
|
8UP1
 
 | |
8UTM
 
 | |
6YVP
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) complexed with dGMP and refined to 2.77 A | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Krakowiak, A, Nawrot, B.C. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
8GLV
 
 | |
5NNQ
 
 | |
6BRU
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a (S)-2-phenylaminopteridinone inhibitor | | Descriptor: | (7S)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7,8-trimethyl-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Aquino, B, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a (S)-2-phenylaminopteridinone inhibitor
To Be Published
|
|
2VUX
 
 | | Human ribonucleotide reductase, subunit M2 B | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE SUBUNIT M2 B | | Authors: | Welin, M, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, van Den Berg, S, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Ribonucleotide Reductase, Subunit M2 B
To be Published
|
|
6Z2Q
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (GalGalNAc-TS) product | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Glycodrosocin, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
2PAS
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Padilla, A, Cave, A, Parello, J, Etienne, G, Baldellon, C. | | Deposit date: | 1994-03-22 | | Release date: | 1994-06-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin
To be Published
|
|
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4CFN
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-(cyclohexylmethoxy)-8-(trifluoromethyl)-9H-purin-2-amine, CYCLIN-A2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
6BNF
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, mono-zinc complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
2VXJ
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH AGAL13BGAL14GLC AT 1.9 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PA-I GALACTOPHILIC LECTIN, ... | | Authors: | Blanchard, B, Nurisso, A, Hollville, E, Tetaud, C, Wiels, J, Pokorna, M, Wimmerova, M, Varrot, A, Imberty, A. | | Deposit date: | 2008-07-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of the Preferential Binding for Globo-Series Glycosphingolipids Displayed by Pseudomonas Aeruginosa Lectin I.
J.Mol.Biol., 383, 2008
|
|
4C13
 
 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
4LQ6
 
 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6BV9
 
 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana after overnight soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, CHLORIDE ION, Proteinaceous RNase P 1, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
6FQF
 
 | | THE X-RAY STRUCTURE OF FERRIC-BETA4 HUMAN HEMOGLOBIN | | Descriptor: | Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Merlino, A, Balasco, N, Balsamo, A, Vergara, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ferric homotetrameric beta4human hemoglobin.
Biophys. Chem., 240, 2018
|
|
3ZJU
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3016 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
1YSP
 
 | | Crystal structure of the C-terminal domain of E. coli transcriptional regulator KdgR. | | Descriptor: | SULFATE ION, Transcriptional regulator kdgR | | Authors: | Bochkarev, A, Lunin, V.V, Ezersky, A, Evdokimova, E, Skarina, T, Xu, X, Borek, D, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of effector binding specificity in IclR transcriptional regulators
To be Published
|
|
