8AJJ
 
 | |
6O15
 
 | | Crystal structure of a putative oxidoreductase YjhC from Escherichia coli in complex with NAD(H) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, Uncharacterized oxidoreductase YjhC | | Authors: | Horne, C.R, Kind, L, Davies, J.S, Dobson, R.C. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | On the structure and function of Escherichia coli YjhC: An oxidoreductase involved in bacterial sialic acid metabolism.
Proteins, 88, 2020
|
|
8AJK
 
 | | Crystal structure of a C43S variant from the disulfide reductase MerA from Staphylococcus aureus | | Descriptor: | FAD-containing oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Weiland, P, Altegoer, F, Bange, G. | | Deposit date: | 2022-07-28 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MerA functions as a hypothiocyanous acid reductase and defense mechanism in Staphylococcus aureus.
Mol.Microbiol., 119, 2023
|
|
5E4E
 
 | | Engineered Interleukin-13 bound to receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-13, Interleukin-13 receptor subunit alpha-1, ... | | Authors: | Moraga, I, Thomas, C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Instructive roles for cytokine-receptor binding parameters in determining signaling and functional potency.
Sci.Signal., 8, 2015
|
|
8EC4
 
 | |
8EBI
 
 | |
8ECF
 
 | |
8EBR
 
 | |
8BVB
 
 | |
6B1I
 
 | |
3VDQ
 
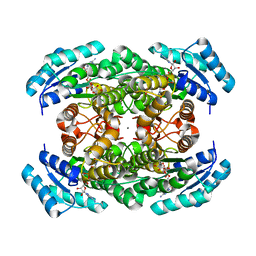 | | Crystal structure of alcaligenes faecalis D-3-hydroxybutyrate dehydrogenase in complex with NAD(+) and acetate | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hoque, M.M, Shimizu, S, Hossain, M.T, Yamamoto, T, Suzuki, K, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of Alcaligenes faecalis D-3-hydroxybutyrate dehydrogenase before and after NAD+ and acetate binding suggest a dynamical reaction mechanism as a member of the SDR family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6EUD
 
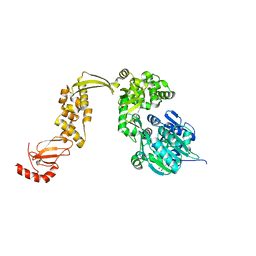 | |
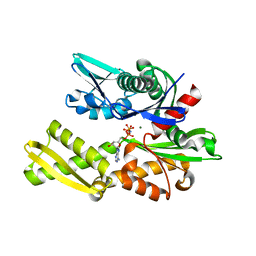
6B1M
 
 | |
6F0E
 
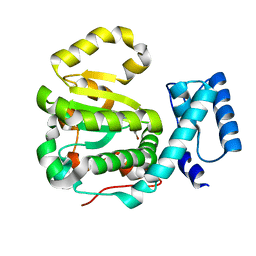 | | Structure of yeast Sec14p with a picolinamide compound | | Descriptor: | SEC14 cytosolic factor, ~{N}-(1,3-benzodioxol-5-ylmethyl)-5-bromanyl-3-fluoranyl-pyridine-2-carboxamide | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties.
Cell Chem Biol, 25, 2018
|
|
6CMG
 
 | |
6CIP
 
 | |
6CIN
 
 | |
6B1N
 
 | |
6CIO
 
 | | Pyruvate:ferredoxin oxidoreductase from Moorella thermoacetica with lactyl-TPP bound | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Chen, P.Y.-T, Drennan, C.L. | | Deposit date: | 2018-02-24 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Binding site for coenzyme A revealed in the structure of pyruvate:ferredoxin oxidoreductase fromMoorella thermoacetica.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CMI
 
 | |
6CIQ
 
 | |
4A5M
 
 | | Redox regulator HypR in its oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UNCHARACTERIZED HTH-TYPE TRANSCRIPTIONAL REGULATOR YYBR | | Authors: | Palm, G.J, Waack, P, Read, R.J, Hinrichs, W. | | Deposit date: | 2011-10-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights Into the Redox-Switch Mechanism of the Marr/Duf24-Type Regulator Hypr
Nucleic Acids Res., 40, 2012
|
|
4A5N
 
 | | Redoxregulator HypR in its reduced form | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, UNCHARACTERIZED HTH-TYPE TRANSCRIPTIONAL REGULATOR YYBR | | Authors: | Palm, G.J, Waack, P, Read, R.J, Hinrichs, W. | | Deposit date: | 2011-10-26 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Insights Into the Redox-Switch Mechanism of the Marr/Duf24-Type Regulator Hypr
Nucleic Acids Res., 40, 2012
|
|
7VRM
 
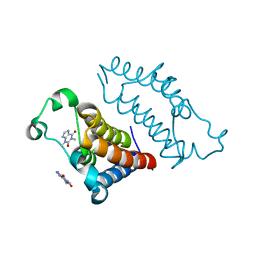 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, THEOPHYLLINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRO
 
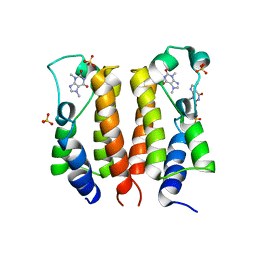 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, SULFATE ION, THEOBROMINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
