3WY7
 
 | | Crystal structure of Mycobacterium smegmatis 7-Keto-8-aminopelargonic acid (KAPA) synthase BioF | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Fan, S.H, Li, D.F, Wang, D.C, Chen, G.J, Zhang, X.E, Bi, L.J. | | Deposit date: | 2014-08-20 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase
Int.J.Biochem.Cell Biol., 58C, 2014
|
|
7C83
 
 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-05-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
5YC7
 
 | |
7CUJ
 
 | | Crystal structure of fission yeast Ccq1 and Tpz1 | | Descriptor: | Coiled-coil quantitatively-enriched protein 1, Protection of telomeres protein tpz1 | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUH
 
 | | Crystal structure of fission yeast Pot1 and ssDNA | | Descriptor: | Protection of telomeres protein 1, Telomere single-strand DNA | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUI
 
 | | Crystal structure of fission yeast Pot1 and Tpz1 | | Descriptor: | Protection of telomeres protein 1, Protection of telomeres protein tpz1, SULFATE ION | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7WZO
 
 | |
5ZUG
 
 | | Structure of the bacterial acetate channel SatP | | Descriptor: | Succinate-acetate/proton symporter SatP, nonyl beta-D-glucopyranoside | | Authors: | Sun, P.C, Li, J.L, Xiao, Q.J, Guan, Z.Y, Deng, D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel.
J. Biol. Chem., 293, 2018
|
|
7DMY
 
 | | The crystal structure of Cpd7 in complex with BPTF bromodomain | | Descriptor: | Nucleosome-remodeling factor subunit BPTF, tert-butyl 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-4-oxidanylidene-5,7-dihydropyrrolo[3,4-d]pyrimidine-6-carboxylate | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7DN4
 
 | | The crystal structure of Cpd8 in complex with BPTF bromodomain | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
8ID1
 
 | |
8IKH
 
 | | Cryo-EM structure of human receptor with G proteins | | Descriptor: | 3-[(1R)-1-(2-methoxyphenyl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, S.Y, Shao, Z.H. | | Deposit date: | 2023-02-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8IKG
 
 | | Cryo-EM structure of human receptor with G proteins | | Descriptor: | 3-[(1S)-1-(furan-2-yl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, S.Y, Shao, Z.H. | | Deposit date: | 2023-02-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ID0
 
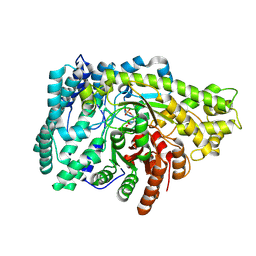 | | Crystal structure of PflD bound to 1,5-anhydromannitol-6-phosphate in Streptococcus dysgalactiae subsp. equisimilis | | Descriptor: | [(2R,3S,4R,5R)-3,4,5-tris(oxidanyl)oxan-2-yl]methyl dihydrogen phosphate, formate C-acetyltransferase | | Authors: | Ma, K.L, Zhang, Y. | | Deposit date: | 2023-02-11 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8ID7
 
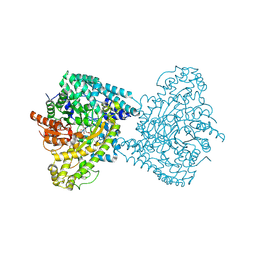 | |
8J5X
 
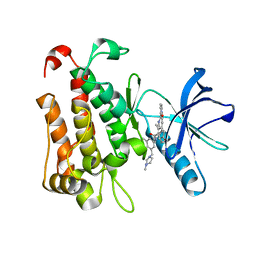 | | The crystal structure of TrkA(G595R) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09192252 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J5W
 
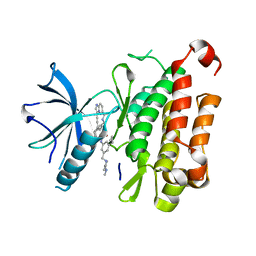 | | The crystal structure of TrkA(F589L) kinase in complex with N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | High affinity nerve growth factor receptor, N-(3-cyclopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-4^6-methyl-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28041458 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
7EKI
 
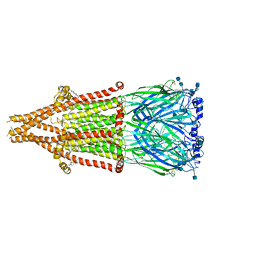 | | human alpha 7 nicotinic acetylcholine receptor in apo-form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKP
 
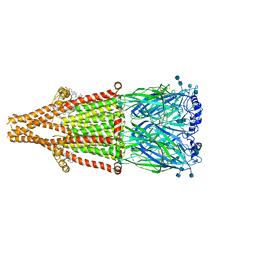 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
7EKT
 
 | | human alpha 7 nicotinic acetylcholine receptor bound to EVP-6124 and PNU-120596 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-chloro-N-(quinuclidin-3-yl)benzo[b]thiophene-2-carboxamide, ... | | Authors: | Liu, S, Zhao, Y, Sun, D, Tian, C. | | Deposit date: | 2021-04-06 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of human alpha 7 nicotinic acetylcholine receptor activation.
Cell Res., 31, 2021
|
|
8J61
 
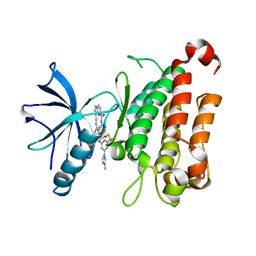 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-14-oxo-5-oxa-13-aza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05065274 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
8J63
 
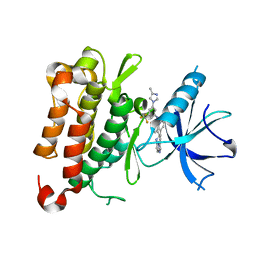 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.0005 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
5Z1C
 
 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
8IOF
 
 | |
6BE0
 
 | | AvrA delL154 with IP6, CoA | | Descriptor: | AvrA, COENZYME A, INOSITOL HEXAKISPHOSPHATE | | Authors: | Labriola, J.M, Nagar, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural Analysis of the Bacterial Effector AvrA Identifies a Critical Helix Involved in Substrate Recognition.
Biochemistry, 57, 2018
|
|
