5Y9F
 
 | |
5Y9C
 
 | |
5Y9E
 
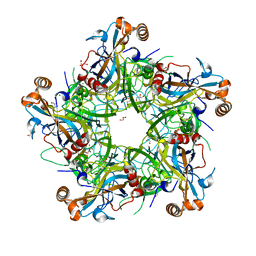 | | Crystal structure of HPV58 pentamer | | Descriptor: | GLYCEROL, MAGNESIUM ION, Major capsid protein L1 | | Authors: | Li, S.W, Li, Z.H. | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.
MBio, 8, 2017
|
|
7ALL
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT4683-S1_4) | | Descriptor: | Arylsulfatase, CALCIUM ION, IODIDE ION, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Martens, E.C, Basle, A, Cartmell, A. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7AN1
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | Arylsulfatase, CALCIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7ANB
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7ANA
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7WUX
 
 | | Crystal structure of AziU3/U2 complexed with (5S,6S)-O7-sulfo DADH from Streptomyces sahachiroi | | Descriptor: | (2S,5S,6S)-2,6-bis(azanyl)-5-oxidanyl-7-sulfooxy-heptanoic acid, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, AziU2, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
7WUW
 
 | | Crystal structure of AziU3/U2 from Streptomyces sahachiroi | | Descriptor: | AziU2, AziU3, MAGNESIUM ION, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
4FFY
 
 | | Crystal structure of DENV1-E111 single chain variable fragment bound to DENV-1 DIII, strain 16007. | | Descriptor: | CHLORIDE ION, DENV1-E111 single chain variable fragment (heavy chain), DENV1-E111 single chain variable fragment (light chain), ... | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
4FFZ
 
 | | Crystal Structure of DENV1-E111 fab fragment bound to DENV-1 DIII (Western Pacific-74 strain). | | Descriptor: | DENV1-E111 fab fragment (heavy chain), DENV1-E111 fab fragment (light chain), Envelope protein E | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
5J8C
 
 | | Human MOF C316S, E350Q crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, SODIUM ION, ... | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
5J8F
 
 | | Human MOF K274P crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, ZINC ION | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
8BVD
 
 | | FimH lectin domain in complex with mannose C-linked to quinoline | | Descriptor: | (2R,3S,4R,5S,6R)-2-(hydroxymethyl)-6-[(E)-3-quinolin-6-ylprop-2-enyl]oxane-3,4,5-triol, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Bouckaert, J, Bridot, C. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Insightful Improvement in the Design of Potent Uropathogenic E. coli FimH Antagonists.
Pharmaceutics, 15, 2023
|
|
4GIH
 
 | |
4GFM
 
 | | JAK2 kinase (JH1 domain) with 2,6-DICHLORO-N-(2-OXO-2,5-DIHYDROPYRIDIN-4-YL)BENZAMIDE | | Descriptor: | 2,6-dichloro-N-(2-oxo-2,5-dihydropyridin-4-yl)benzamide, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
4GMY
 
 | |
6FH5
 
 | | PI3Kg IN COMPLEX WITH Compound 7 | | Descriptor: | 3-methyl-1-(oxan-4-yl)-8-pyridin-3-yl-imidazo[4,5-c]quinolin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Barlaam, B. | | Deposit date: | 2018-01-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery and Optimisation of a Novel Series of 3-Cinnoline Carboxamides as Orally Bioavailable, Highly Potent and Selective Ataxia Telangiectasia Mutated (ATM) inhibitors
To Be Published
|
|
7BJ6
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-3-(4-chlorophenyl)-4-fluoranyl-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BIT
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-3-(4-chlorophenyl)-4-fluoranyl-3-[(1-oxidanylcyclopropyl)methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BIV
 
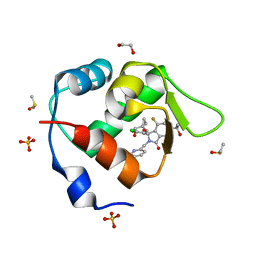 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1,2-ETHANEDIOL, 6-[[(1~{R})-1-(4-chlorophenyl)-7-fluoranyl-1-[[1-(hydroxymethyl)cyclopropyl]methoxy]-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-2-yl]methyl]pyridine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BJ0
 
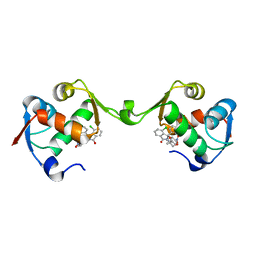 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-4-chloranyl-3-(4-chlorophenyl)-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-2-[(4-nitrophenyl)methyl]isoindol-1-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BMG
 
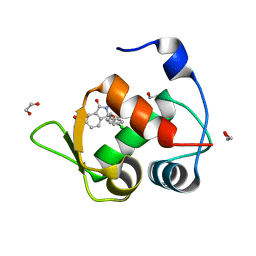 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-3-(4-chlorophenyl)-2-[(4-ethynylphenyl)methyl]-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-20 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BIR
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1-[[(1~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-1-(4-chlorophenyl)-7-fluoranyl-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-1-yl]oxymethyl]cyclopropane-1-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6R4N
 
 | | Crystal structure of S. cerevisia Niemann-Pick type C protein NPC2 with ergosterol bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ERGOSTEROL, ... | | Authors: | Winkler, M.B.L, Kidmose, R.T, Pedersen, B.P. | | Deposit date: | 2019-03-22 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight into Eukaryotic Sterol Transport through Niemann-Pick Type C Proteins.
Cell, 179, 2019
|
|
