7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
6KLH
 
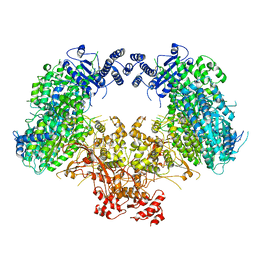 | | Dimeric structure of Machupo virus polymerase bound to vRNA promoter | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6KLC
 
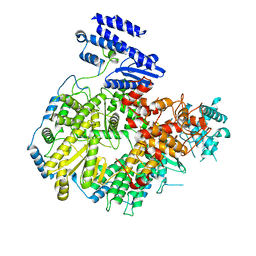 | | Structure of apo Lassa virus polymerase | | Descriptor: | MANGANESE (II) ION, RNA-directed RNA polymerase L | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
6KLE
 
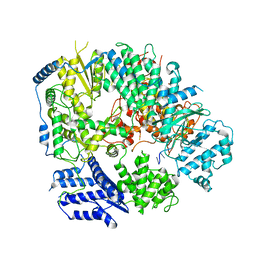 | | Monomeric structure of Machupo virus polymerase bound to vRNA promoter | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*GP*CP*CP*UP*AP*GP*GP*AP*UP*CP*CP*AP*CP*UP*GP*UP*GP*CP*G)-3'), RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
6KLD
 
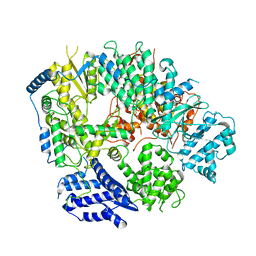 | | Structure of apo Machupo virus polymerase | | Descriptor: | MANGANESE (II) ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Peng, R, Xu, X, Jing, J, Peng, Q, Gao, G.F, Shi, Y. | | Deposit date: | 2019-07-30 | | Release date: | 2020-03-18 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insight into arenavirus replication machinery.
Nature, 579, 2020
|
|
5ZQ2
 
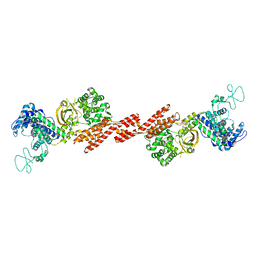 | | SidE apo form | | Descriptor: | SidE | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
6L5Z
 
 | |
6LJM
 
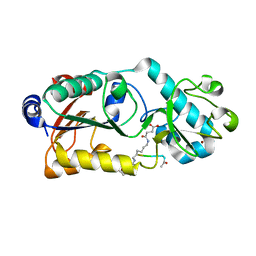 | |
6L4Z
 
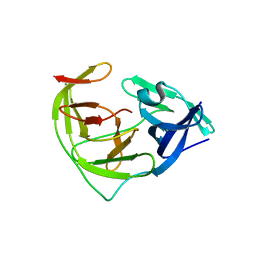 | | Crystal structure of Zika NS2B-NS3 protease with compound 6 | | Descriptor: | 4-(hydroxymethyl)benzoic acid, Genome polyprotein | | Authors: | Quek, J.P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
6LJN
 
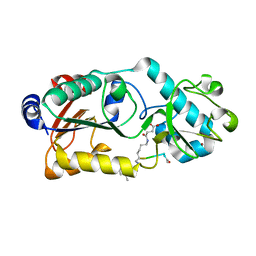 | |
6L50
 
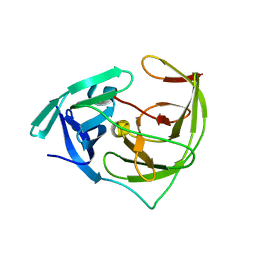 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | Descriptor: | 2-sulfanylidene-1,3-thiazolidin-4-one, NS3 protease, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
5ZQ3
 
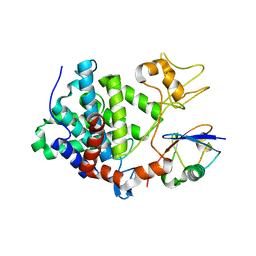 | | PDE-Ubiquitin | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ5
 
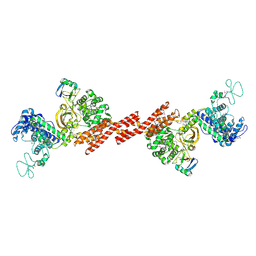 | | SidE-Ubi | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
6LJK
 
 | |
5YX2
 
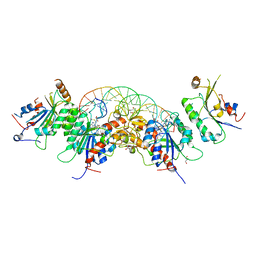 | |
5ZQ4
 
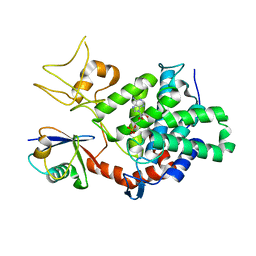 | | PDE-Ubi-ADPr | | Descriptor: | ADENOSINE MONOPHOSPHATE, SidE, ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
6BFG
 
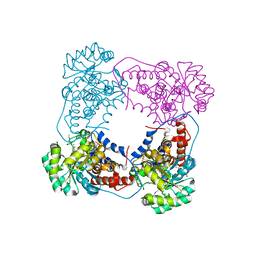 | |
7Y67
 
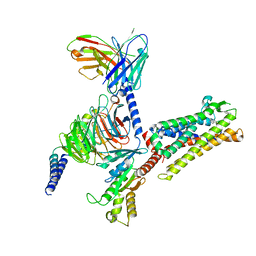 | | Cryo-EM structure of C089-bound C5aR1(I116A) mutant in complex with Gi protein | | Descriptor: | C089 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y65
 
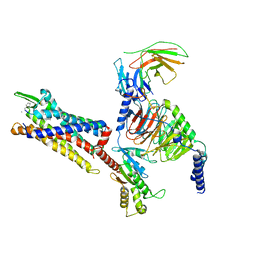 | | Cryo-EM structure of C5a peptide-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, C5apep peptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y64
 
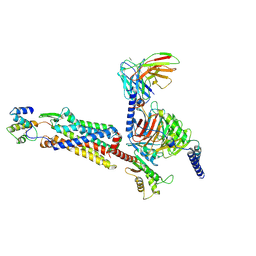 | | Cryo-EM structure of C5a-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y66
 
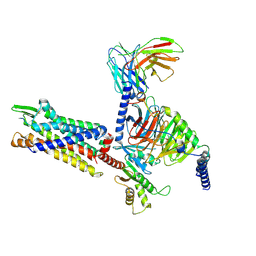 | | Cryo-EM structure of BM213-bound C5aR1 in complex with Gi protein | | Descriptor: | BM213 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
5ZQ7
 
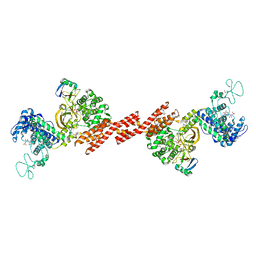 | | SidE-Ubi-NAD | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SidE, ... | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
