6WAV
 
 | | Crystal structure of PHF1 in complex with H3K36me3 substitution | | Descriptor: | Histone H3.1, PHD finger protein 1, SULFATE ION, ... | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6WAT
 
 | | complex structure of PHF1 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 1, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
5LF9
 
 | | Crystal structure of human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22 | | Authors: | Tallant, C, Siejka, P, Mathea, S, Shrestha, L, Krojer, T, Srikannathasan, V, Elkins, J.M, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human NUDT22
To Be Published
|
|
5JUW
 
 | | complex of Dot1l with SS148 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Li, Y, Spurr, S.S, Bayle, E.D, Fish, P.V, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complex of Dot1l with SS148
To Be Published
|
|
6P9G
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 2-(4-oxoquinazolin-3(4H)-yl)propanoic acid | | Descriptor: | (2R)-2-(4-oxoquinazolin-3(4H)-yl)propanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Tempel, W, Mann, M.K, Harding, R.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
6YG1
 
 | | Crystal structure of MKK7 (MAP2K7) in an active state, allosterically triggered by the N-terminal helix | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, SODIUM ION | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC), Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
5LF8
 
 | | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17) | | Descriptor: | ETHYL MERCURY ION, Nucleoside diphosphate-linked moiety X motif 17, PHOSPHATE ION | | Authors: | Mathea, S, Tallant, C, Salah, E, Wang, D, Velupillai, S, Nowak, R, Oerum, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17)
To Be Published
|
|
4QQD
 
 | | Crystal Structure of tandem tudor domains of UHRF1 in complex with a small organic molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, E3 ubiquitin-protein ligase UHRF1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Iqbal, A, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
6PED
 
 | | Crystal structure of HEMK2-TRMT112 complex | | Descriptor: | Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HEMK2-TRMT112 complex
To Be Published
|
|
6PDM
 
 | | Crystal structure of Human Protein Arginine Methyltransferase 9 (PRMT9) | | Descriptor: | Protein arginine N-methyltransferase 9, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Tempel, W, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Human Protein Arginine Methyltransferase 9 (PRMT9)
To Be Published
|
|
2QKX
 
 | | N-acetyl glucosamine 1-phosphate uridyltransferase from Mycobacterium tuberculosis complex with N-acetyl glucosamine 1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein glmU | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-07-11 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of GlmU from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7PC6
 
 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
6YFZ
 
 | |
6YG3
 
 | | Crystal structure of MKK7 (MAP2K7) covalently bound with CPT1-70-1 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, N-(4-((2-((4-(4-methylpiperazin-1-yl)phenyl)amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy)phenyl)acrylamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6YG7
 
 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor SB1-G-23 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{R})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6Y24
 
 | | Crystal structure of fourth KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1 | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6YG4
 
 | |
6Q94
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
6YG5
 
 | |
6YG0
 
 | |
6YG6
 
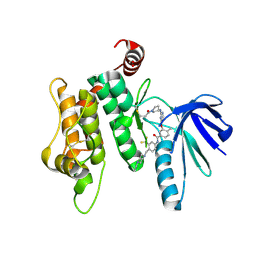 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor TL10-105 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{S})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6QB7
 
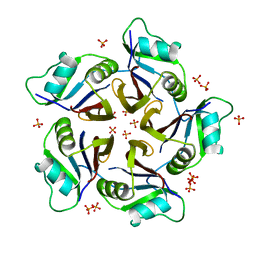 | | Structure of the H1 domain of human KCTD16 | | Descriptor: | BTB/POZ domain-containing protein KCTD16, PHOSPHATE ION | | Authors: | Pinkas, D.M, Bufton, J.C, Williams, E.P, Strain-Damerell, C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-20 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the H1 domain of human KCTD16
To be published
|
|
6Y7H
 
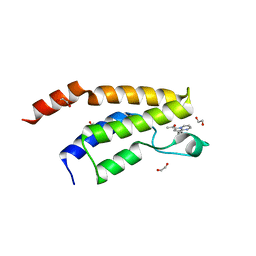 | | Structure of the BRD9 bromodomain and compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-phenylpyrrolo[1,2-a]pyrimidin-6-yl)ethanone, Bromodomain-containing protein 9 | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the BRD9 bromodomain
To Be Published
|
|
6Y7G
 
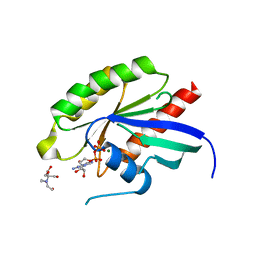 | | Structure of the human RAB3C in complex with GDP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Jung, S, Raux, B, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-29 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human RAB3C in complex with GDP
To Be Published
|
|
6Y7I
 
 | | Structure of the BRD9 bromodomain and compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-pyridin-2-ylpyrrolo[1,2-a]pyrimidin-6-yl)ethanone, Bromodomain-containing protein 9 | | Authors: | Diaz-Saez, L, Krojer, T, Picaud, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-01 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the BRD9 bromodomain and compound 2
To Be Published
|
|
