5FD6
 
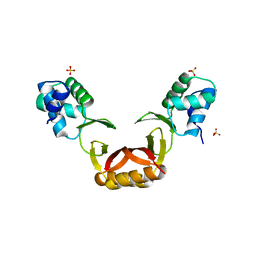 | |
5FSR
 
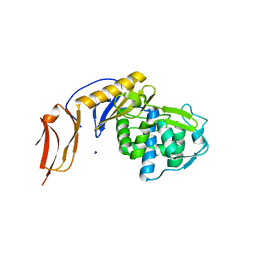 | | Crystal structure of penicillin binding protein 6B from Escherichia coli | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE DACD | | Authors: | Peters, K, Kannan, S, Rao, V.A, Bilboy, J, Vollmer, D, Erickson, S.W, Lewis, R.J, Young, K.D, Vollmer, W. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia Coli
Mbio, 7, 2016
|
|
5FMV
 
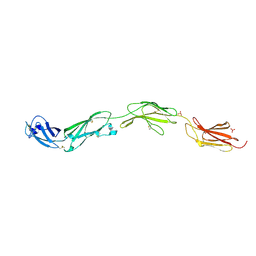 | | Crystal structure of human CD45 extracellular region, domains d1-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C, SULFATE ION | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FUW
 
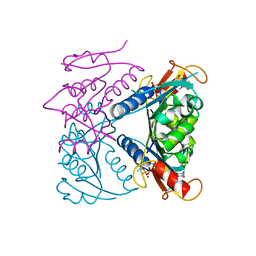 | | catalytic domain of Thymidine kinase from Trypanosoma brucei with dTMP or dThd | | Descriptor: | PHOSPHATE ION, THYMDINE KINASE, THYMIDINE, ... | | Authors: | Timm, J, Valente, M, Castillo-Acosta, V, Balzarini, T, Nettleship, J.E, Rada, H, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2016-01-31 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cell Cycle Regulation and Novel Structural Features of Thymidine Kinase, an Essential Enzyme in Trypanosoma Brucei.
Mol.Microbiol., 102, 2016
|
|
5FN6
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN8
 
 | | Crystal structure of rat CD45 extracellular region, domains d3-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FQL
 
 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | Deposit date: | 2015-12-11 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
1AK0
 
 | | P1 NUCLEASE IN COMPLEX WITH A SUBSTRATE ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-(DITHIO)PHOSPHATE, ... | | Authors: | Romier, C, Suck, D. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs.
Proteins, 32, 1998
|
|
5FZP
 
 | | Structure of the dispase autolysis inducing protein from Streptomyces mobaraensis | | Descriptor: | CALCIUM ION, DISPASE AUTOLYSIS-INDUCING PROTEIN, GLYCEROL | | Authors: | Schmelz, S, Fiebig, D, Beck, J, Fuchsbauer, H.L, Scrima, A. | | Deposit date: | 2016-03-15 | | Release date: | 2016-08-10 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Dispase Autolysis Inducing Protein from Streptomyces Mobaraensis and Glutamine Cross-Linking Sites for Transglutaminase
J.Biol.Chem., 291, 2016
|
|
5G04
 
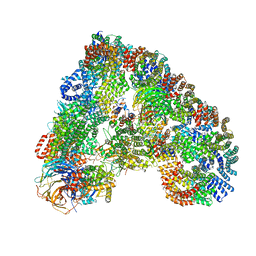 | | Structure of the human APC-Cdc20-Hsl1 complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Zhang, S, Chang, L, Alfieri, C, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Mechanism of Apc/C Activation by Mitotic Phosphorylation.
Nature, 533, 2016
|
|
5G0F
 
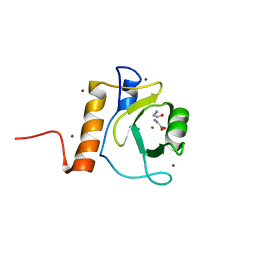 | | Crystal structure of Danio rerio HDAC6 ZnF-UBP domain | | Descriptor: | GLYCINE, HDAC6, NICKEL (II) ION, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
5FNT
 
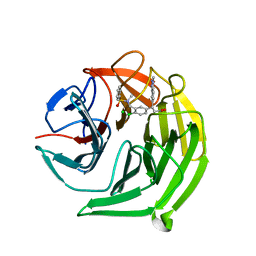 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
8RF9
 
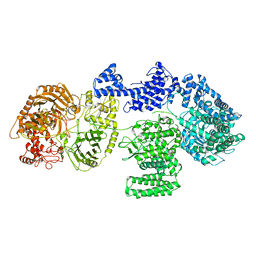 | | CgsiGP1 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
5FZJ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
3SGS
 
 | |
7K4P
 
 | | Crystal structure of Kemp Eliminase HG3 | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
3OE1
 
 | | Pyruvate decarboxylase variant Glu473Asp from Z. mobilis in complex with reaction intermediate 2-lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Meyer, D, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Double duty for a conserved glutamate in pyruvate decarboxylase: evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate .
Biochemistry, 49, 2010
|
|
7K4S
 
 | | Crystal structure of Kemp Eliminase HG3.7 | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
3ZQH
 
 | | Structure of Tetracycline repressor in complex with inducer peptide- TIP3 | | Descriptor: | 1,2-ETHANEDIOL, INDUCER PEPTIDE TIP3, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, ... | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
1AHO
 
 | | THE AB INITIO STRUCTURE DETERMINATION AND REFINEMENT OF A SCORPION PROTEIN TOXIN | | Descriptor: | TOXIN II | | Authors: | Smith, G.D, Blessing, R.H, Ealick, S.E, Fontecilla-Camps, J.C, Hauptman, H.A, Housset, D, Langs, D.A, Miller, R. | | Deposit date: | 1997-04-08 | | Release date: | 1997-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Ab initio structure determination and refinement of a scorpion protein toxin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7K4Y
 
 | | Crystal structure of Kemp Eliminase HG3.17 at 343 K | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5ET5
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET8
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | T-to-R switch of muscle FBPase involves extreme changes of secondary and quaternary structure
To Be Published
|
|
5ET6
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET7
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
