4L23
 
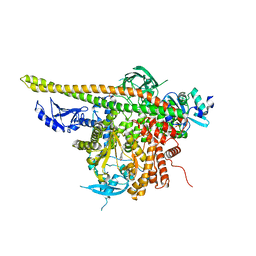 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4L2Y
 
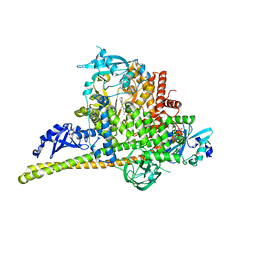 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and compound 9d | | Descriptor: | 3-amino-5-[4-(morpholin-4-yl)pyrido[3',2':4,5]furo[3,2-d]pyrimidin-2-yl]phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4L1B
 
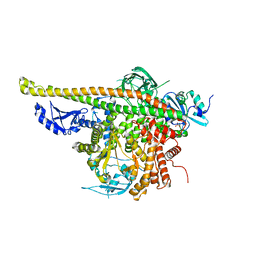 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
6A82
 
 | |
6A83
 
 | |
6IWT
 
 | |
8VPN
 
 | | Phosphorylated human NCC in complex with indapamide | | Descriptor: | 4-chloro-N-[(2S)-2-methyl-2,3-dihydro-1H-indol-1-yl]-3-sulfamoylbenzamide, ADENOSINE-5'-TRIPHOSPHATE, Solute carrier family 12 member 3 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2024-01-16 | | Release date: | 2024-09-04 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural bases for Na + -Cl - cotransporter inhibition by thiazide diuretic drugs and activation by kinases.
Nat Commun, 15, 2024
|
|
8VPP
 
 | | Phosphorylated human NCC in complex with chlorthalidone | | Descriptor: | 2-chloro-5-[(1S)-1-hydroxy-3-oxo-2H-isoindol-1-yl]benzenesulfonamide, Solute carrier family 12 member 3 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2024-01-16 | | Release date: | 2024-09-04 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural bases for Na + -Cl - cotransporter inhibition by thiazide diuretic drugs and activation by kinases.
Nat Commun, 15, 2024
|
|
1VBS
 
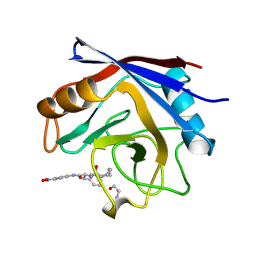 | | STRUCTURE OF CYCLOPHILIN COMPLEXED WITH (D)ALA CONTAINING TETRAPEPTIDE | | Descriptor: | CYCLOPHILIN A, TETRAPEPTIDE | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the stereospecificity of peptidyl prolyl cis/trans isomerases.
FEBS Lett., 432, 1998
|
|
8EG6
 
 | | huCaspase-6 in complex with inhibitor 2a | | Descriptor: | (3R)-1-(ethanesulfonyl)-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
8EG5
 
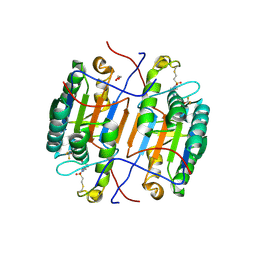 | | huCaspase-6 in complex with inhibitor 3a | | Descriptor: | (3R,5S)-1-(ethanesulfonyl)-5-phenyl-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide (bound form), 1,2-ETHANEDIOL, Caspase-6 subunit p11, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
7C72
 
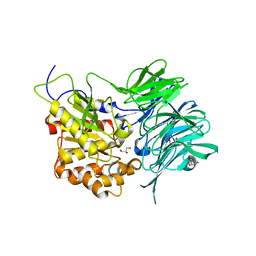 | | Structure of a mycobacterium tuberculosis puromycin-hydrolyzing peptidase | | Descriptor: | D-MALATE, GLYCEROL, Prolyl oligopeptidase | | Authors: | Ruiz-Carrillo, D, Zhao, Y.H, Feng, Q, Zhou, X, Zhang, Y, Jiang, J, Lukman, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.00004458 Å) | | Cite: | Mycobacterium tuberculosis puromycin hydrolase displays a prolyl oligopeptidase fold and an acyl aminopeptidase activity.
Proteins, 89, 2021
|
|
8J9Z
 
 | |
8J9Y
 
 | |
7X4B
 
 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7CIR
 
 | | Peptide phosphorylation modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIQ
 
 | | Phosphorylation modification of MHC I polypeptide | | Descriptor: | ARG-ARG-PHE-SER-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIS
 
 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7DYN
 
 | | Phosphorylation of MHC I peptide | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
8CEG
 
 | | BAR domain protein FAM92A1 essential for mitochondrial membrane remodeling | | Descriptor: | CBY1-interacting BAR domain-containing protein 1 | | Authors: | Kajander, T, Fudo, S, Yan, Z, Zhao, H. | | Deposit date: | 2023-02-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Membrane remodeling by FAM92A1 during brain development regulates neuronal morphology, synaptic function, and cognition.
Nat Commun, 15, 2024
|
|
5QIN
 
 | |
5QIM
 
 | |
5QIL
 
 | |
7NX6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
