1GGD
 
 | | CRYSTAL STRUCTURE OF GAMMA CHYMOTRYPSIN WITH N-ACETYL-LEUCIL-PHENYLALANINE ALDEHYDE BOUND AT THE ACTIVE SITE | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID (1-FORMYL-2-PHENYL-ETHYL)-AMIDE, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Neidhart, D, Wei, Y, Cassidy, C, Lin, J, Cleland, W.W, Frey, P.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin.
Biochemistry, 40, 2000
|
|
6L62
 
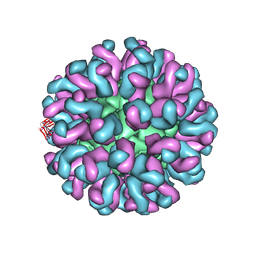 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
8YJN
 
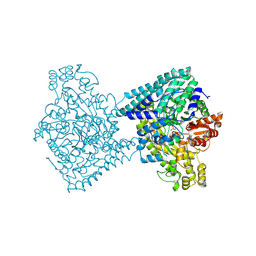 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8YJO
 
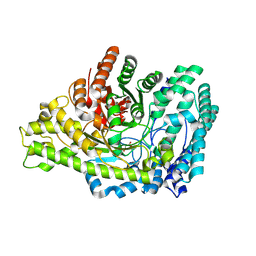 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
1JXZ
 
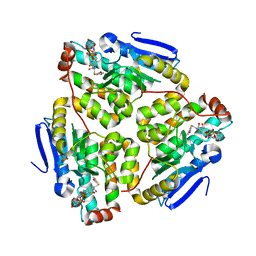 | | Structure of the H90Q mutant of 4-Chlorobenzoyl-Coenzyme A Dehalogenase complexed with 4-hydroxybenzoyl-Coenzyme A (product) | | Descriptor: | 4-HYDROXYBENZOYL COENZYME A, 4-chlorobenzoyl Coenzyme A dehalogenase, CALCIUM ION, ... | | Authors: | Thoden, J.B, Zhang, W, Wei, Y, Luo, L, Taylor, K.L, Yang, G, Dunaway-Mariano, D, Benning, M.M, Holden, H.M. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 90 Function in 4-chlorobenzoyl-coenzyme A Dehalogenase Catalysis
Biochemistry, 40, 2001
|
|
1TLH
 
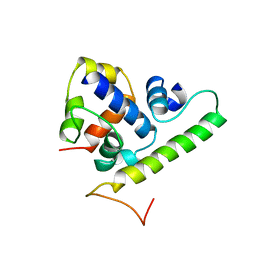 | | T4 AsiA bound to sigma70 region 4 | | Descriptor: | 10 kDa anti-sigma factor, RNA polymerase sigma factor rpoD | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma(70) region 4
Embo J., 23, 2004
|
|
1THT
 
 | | STRUCTURE OF A MYRISTOYL-ACP-SPECIFIC THIOESTERASE FROM VIBRIO HARVEYI | | Descriptor: | THIOESTERASE | | Authors: | Lawson, D.M, Derewenda, U, Serre, L, Ferri, S, Szitter, R, Wei, Y, Meighen, E.A, Derewenda, Z.S. | | Deposit date: | 1994-04-19 | | Release date: | 1995-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi.
Biochemistry, 33, 1994
|
|
1TL6
 
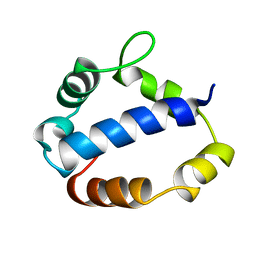 | | Solution structure of T4 bacteriphage AsiA monomer | | Descriptor: | 10 kDa anti-sigma factor | | Authors: | Lambert, L.J, Wei, Y, Schirf, V, Demeler, B, Werner, M.H. | | Deposit date: | 2004-06-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T4 AsiA blocks DNA recognition by remodeling sigma70 region 4
Embo J., 23, 2004
|
|
4EEQ
 
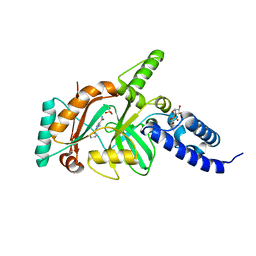 | | Crystal structure of E. faecalis DNA ligase with inhibitor | | Descriptor: | 4-amino-2-(cyclopentyloxy)pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wang, T, Charifson, P, Wei, Y. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis and Activity Evaluation of Potent NAD+ DNA Ligase Inhibitors as Potential Antibacterial Agents.
To be Published
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
6LM3
 
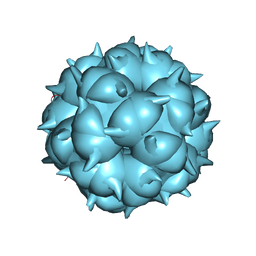 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
2GF5
 
 | | Structure of intact FADD (MORT1) | | Descriptor: | FADD protein | | Authors: | Carrington, P.E, Sandu, C, Wei, Y, Hill, J.M, Morisawa, G, Huang, T, Gavathiotis, E, Wei, Y, Werner, M.H. | | Deposit date: | 2006-03-21 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of FADD and Its Mode of Interaction with Procaspase-8
Mol.Cell, 22, 2006
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
3RSM
 
 | | Crystal structure of S108C mutant of PMM/PGM | | Descriptor: | PHOSPHATE ION, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Akella, A, Anbanandam, A, Kelm, A, Wei, Y, Mehra-Chaudhary, R, Beamer, L, Van Doren, S. | | Deposit date: | 2011-05-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Solution NMR of a 463-residue phosphohexomutase: domain 4 mobility, substates, and phosphoryl transfer defect.
Biochemistry, 51, 2012
|
|
8HGH
 
 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
6JHB
 
 | |
7E88
 
 | |
7Y5N
 
 | | Structure of 1:1 PAPP-A.ProMBP complex(half map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone marrow proteoglycan, ... | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7Y5Q
 
 | | Structure of 1:1 PAPP-A.STC2 complex(half map) | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7XLQ
 
 | | Structure of human R-type voltage-gated CaV2.3-alpha2/delta1-beta1 channel complex in the ligand-free (apo) state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gao, Y, Qiu, Y, Wei, Y, Dong, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-04-22 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into the gating mechanisms of voltage-gated calcium channel Ca V 2.3.
Nat Commun, 14, 2023
|
|
2LS7
 
 | |
7JRS
 
 | |
7JRR
 
 | | Crystal structures of artificially designed homomeric RNA nanoarchitectures | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, RNA (50-MER) | | Authors: | Liu, D, Shao, Y, Piccirilli, J.A, Weizmann, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of artificially designed discrete RNA nanoarchitectures at near-atomic resolution.
Sci Adv, 7, 2021
|
|
