2DNH
 
 | | Solution structure of RNA binding domain in Bruno-like 5 RNA binding protein | | Descriptor: | Bruno-like 5, RNA binding protein | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Bruno-like 5 RNA binding protein
To be Published
|
|
7Y4A
 
 | | Crystal structure of human ELMO1 RBD-RhoG complex | | Descriptor: | Engulfment and cell motility protein 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tsuda, K, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Targeting Ras-binding domain of ELMO1 by computational nanobody design.
Commun Biol, 6, 2023
|
|
6MIB
 
 | |
6JPP
 
 | |
4TV7
 
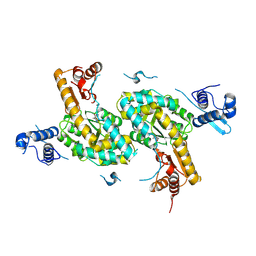 | |
5X7E
 
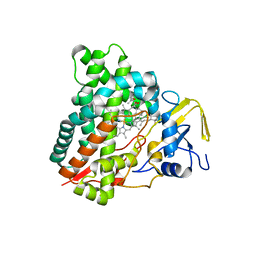 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D2 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(E,2R,5S)-5,6-dimethyl-6-oxidanyl-hept-3-en-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydr o-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D3 dihydroxylase | | Authors: | Hayashi, K, Yasuda, K, Shiro, Y, Sugimoto, H, Sakaki, T. | | Deposit date: | 2017-02-25 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Production of an active form of vitamin D2 by genetically engineered CYP105A1
Biochem. Biophys. Res. Commun., 486, 2017
|
|
7DRU
 
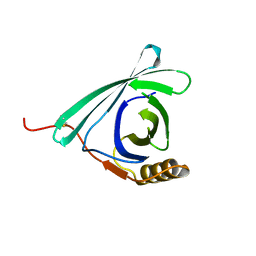 | |
7EWS
 
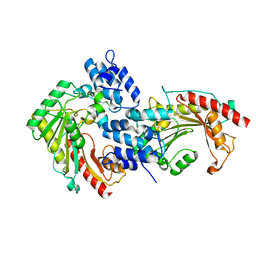 | | Crystal structure of arginine kinase (AK3) from the ciliate Paramecium tetraurelia | | Descriptor: | arginine kinase | | Authors: | Otsuka, Y, Yokota, J, Yano, D, Uda, K, Suzuki, T, Sugiyama, S. | | Deposit date: | 2021-05-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of arginine kinase (AK3) from the ciliate Paramecium tetraurelia
To Be Published
|
|
2RPP
 
 | | Solution structure of Tandem zinc finger domain 12 in Muscleblind-like protein 2 | | Descriptor: | Muscleblind-like protein 2, ZINC ION | | Authors: | Abe, C, Dang, W, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
2RU3
 
 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
1J2A
 
 | | Structure of E. coli cyclophilin B K163T mutant | | Descriptor: | cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2002-12-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
6KB0
 
 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic acid (ETYA) co-crystals obtained by soaking | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB1
 
 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by soaking | | Descriptor: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB8
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAY
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by soaking | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB2
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by soaking | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4D7Y
 
 | | Crystal structure of mouse C1QL1 globular domain | | Descriptor: | C1Q-RELATED FACTOR, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kakegawa, W, Mitakidis, N, Miura, E, Abe, M, Matsuda, K, Takeo, Y, Kohda, K, Motohashi, J, Takahashi, A, Nagao, S, Muramatsu, S, Watanabe, M, Sakimura, K, Aricescu, A.R, Yuzaki, M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Anterograde C1Ql1 Signaling is Required in Order to Determine and Maintain a Single-Winner Climbing Fiber in the Mouse Cerebellum
Neuron, 85, 2015
|
|
2RT9
 
 | | Solution structure of a regulatory domain of meiosis inhibitor | | Descriptor: | F-box only protein 43, ZINC ION | | Authors: | Shoji, S, Muto, Y, Ikeda, M, He, F, Tsuda, K, Ohsawa, N, Akasaka, R, Terada, T, Wakiyama, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation
FEBS Open Bio, 4, 2014
|
|
2RS2
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
2RR6
 
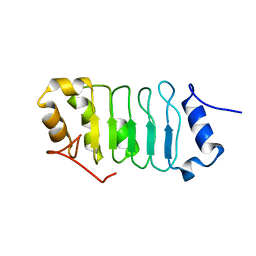 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
2RS7
 
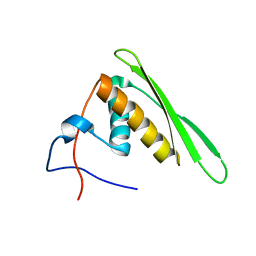 | | Solution structure of the second dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
6LKD
 
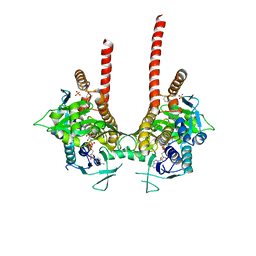 | | in meso full-length rat KMO in complex with a pyrazoyl benzoic acid inhibitor | | Descriptor: | 5-[5-(4-chloranyl-3-fluoranyl-phenyl)-4-methyl-pyrazol-1-yl]-2-phenylmethoxy-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
6LKE
 
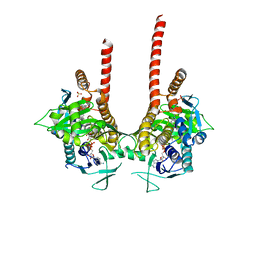 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
2YSO
 
 | | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog | | Descriptor: | ZINC ION, Zinc finger protein 95 homolog | | Authors: | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 656-688) of human Zinc finger protein 95 homolog
To be Published
|
|
