6D1N
 
 | |
6AYH
 
 | | Salmonella enterica GusR | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, GLYCEROL, TetR family transcriptional regulator | | Authors: | Little, M.S, Pellock, S.J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the regulation of beta-glucuronidase expression by human gut Enterobacteriaceae.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6D1P
 
 | | Apo structure of Bacteroides uniformis beta-glucuronidase 3 | | Descriptor: | GLYCEROL, Glycosyl hydrolases family 2, sugar binding domain protein, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D41
 
 | | Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
5DOZ
 
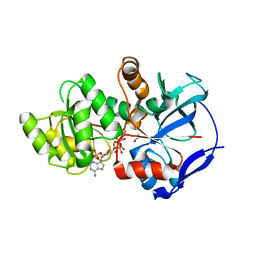 | | Crystal structure of JamJ enoyl reductase (NADPH bound) | | Descriptor: | ACETATE ION, JamJ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
5DOV
 
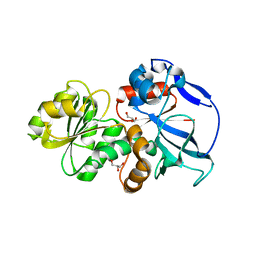 | |
6W32
 
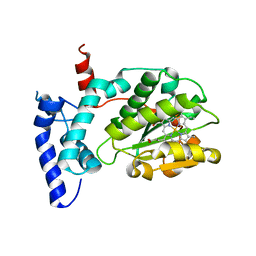 | | Crystal structure of Sfh5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phosphatidylinositol transfer protein SFH5 | | Authors: | Gulten, G, Khan, D, Aggarwal, A, Krieger, I, Sacchettini, J.C, Bankaitis, V.A. | | Deposit date: | 2020-03-08 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Sec14-like phosphatidylinositol transfer protein paralog defines a novel class of heme-binding proteins.
Elife, 9, 2020
|
|
6OD1
 
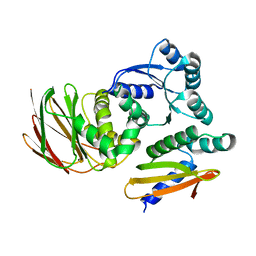 | | IraD-bound to RssB D58P variant | | Descriptor: | Anti-adapter protein IraD, Regulator of RpoS | | Authors: | Deaconescu, A.M, Dorich, V. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of a response regulator of sigmaSstability by a ClpXP antiadaptor.
Genes Dev., 33, 2019
|
|
5DP1
 
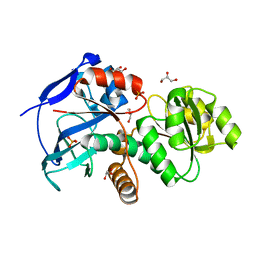 | | Crystal structure of CurK enoyl reductase | | Descriptor: | CurK, GLYCEROL, PHOSPHATE ION | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-12 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
5DP2
 
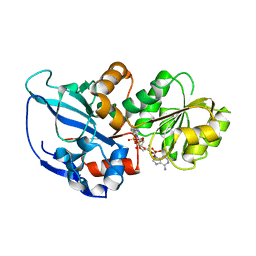 | |
7ZGB
 
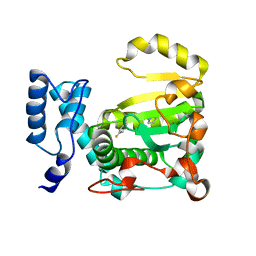 | | Structure of yeast Sec14p with NPPM112 | | Descriptor: | 4-fluoranyl-~{N}-[(4-pyrrolidin-1-ylphenyl)methyl]benzamide, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZG9
 
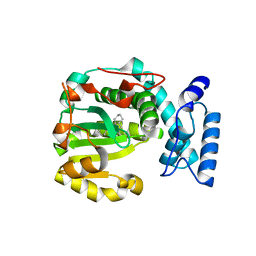 | | Structure of yeast Sec14p with himbacine | | Descriptor: | Himbacine, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGA
 
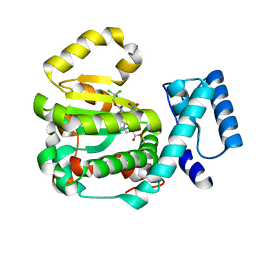 | | Structure of yeast Sec14p with ergoline | | Descriptor: | SEC14 cytosolic factor, ~{O}9-methyl ~{O}4-[2,2,2-tris(chloranyl)ethyl] (5~{a}~{S},6~{a}~{S},9~{R},10~{a}~{S})-7-methyl-3-nitro-5,5~{a},6,6~{a},8,9,10,10~{a}-octahydroindolo[4,3-fg]quinoline-4,9-dicarboxylate | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGC
 
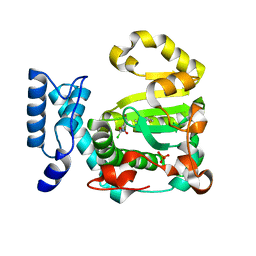 | | Structure of yeast Sec14p with NPPM481 | | Descriptor: | (4-chloranyl-3-nitro-phenyl)-[4-(2-fluorophenyl)piperazin-1-yl]methanone, PHOSPHATE ION, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGD
 
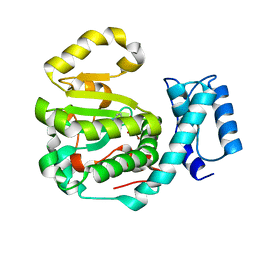 | | Structure of yeast Sec14p with NPPM244 | | Descriptor: | (4-bromanyl-3-nitro-phenyl)-[4-(2-fluorophenyl)piperazin-1-yl]methanone, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
6F0E
 
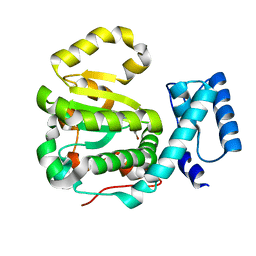 | | Structure of yeast Sec14p with a picolinamide compound | | Descriptor: | SEC14 cytosolic factor, ~{N}-(1,3-benzodioxol-5-ylmethyl)-5-bromanyl-3-fluoranyl-pyridine-2-carboxamide | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties.
Cell Chem Biol, 25, 2018
|
|
4X7U
 
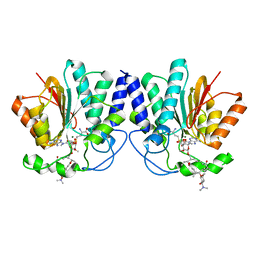 | |
4X81
 
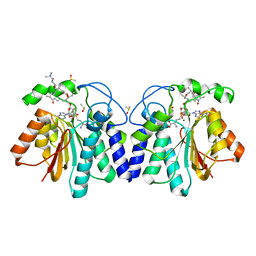 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, M56A, E139A variant) in complex with Mg, SAH and mycinamicin VI (MycE substrate) | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7W
 
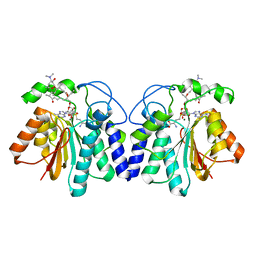 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and mycinamicin VI (MycE substrate) | | Descriptor: | MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, Mycinamicin VI, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7Z
 
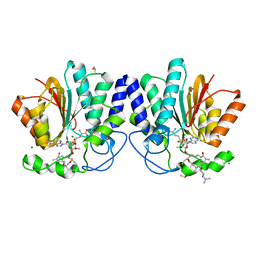 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, M56A, E139A variant) in complex with Mg, SAH and mycinamicin III (substrate) | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, MYCINAMICIN III, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7X
 
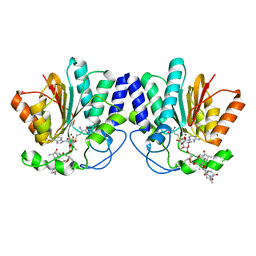 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and macrocin | | Descriptor: | 2-[(4R,5S,6S,7R,9R,11E,13E,15R,16R)-6-[(2R,3R,4R,5S,6R)-4-(dimethylamino)-5-[(2S,4R,5S,6S)-4,6-dimethyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-3-oxidanyl-oxan-2-yl]oxy-16-ethyl-15-[[(2R,3R,4R,5S,6R)-3-methoxy-6-methyl-4,5-bis(oxidanyl)oxan-2-yl]oxymethyl]-5,9,13-trimethyl-4-oxidanyl-2,10-bis(oxidanylidene)-1-oxacyclohexadeca-11,13-dien-7-yl]ethanal, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7Y
 
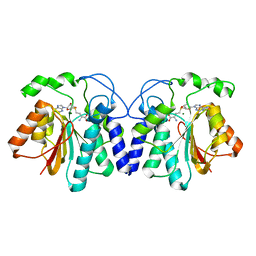 | |
4X7V
 
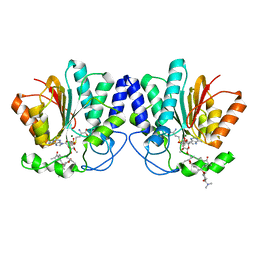 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and mycinamicin IV (product) | | Descriptor: | MAGNESIUM ION, MYCINAMICIN IV, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4XVZ
 
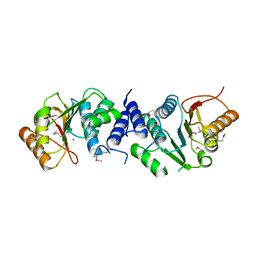 | | MycF mycinamicin III 3'-O-methyltransferase in complex with Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4XVY
 
 | | MycF mycinamicin III 3'-O-methyltransferase in complex with SAH | | Descriptor: | MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
