3GN3
 
 | | Crystal structure of a putative protein-disulfide isomerase from Pseudomonas syringae to 2.5A resolution. | | Descriptor: | GLYCEROL, SULFATE ION, putative protein-disulfide isomerase | | Authors: | Stein, A.J, Chhor, G, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-16 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative protein-disulfide isomerase from Pseudomonas syringae to 2.5A resolution.
To be Published
|
|
3HF7
 
 | | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A | | Descriptor: | ADENOSINE MONOPHOSPHATE, uncharacterized CBS-domain protein | | Authors: | Stein, A.J, Nocek, B, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A
To be Published
|
|
5J6D
 
 | | Discovery of acyl guanidine tryptophan hydroxylase-1 inhibitors | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[(N-{[2-(3-methoxyphenoxy)-6-(piperidin-1-yl)phenyl]methyl}carbamimidoyl)carbamoyl]-L-phenylalanine, FE (III) ION, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of acyl guanidine tryptophan hydroxylase-1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5T31
 
 | | Exploiting an Asp-Glu switch in Glycogen Synthase Kinase 3 to design paralog selective inhibitors for use in acute myeloid leukemia | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Stein, A.J, Holson, E.B, Wagner, F.F, Cambell, A.J. | | Deposit date: | 2016-08-24 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KXA
 
 | | Selective Inhibition of Autotaxin is Effective in Mouse Models of Liver Fibrosis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[6-chloranyl-2-cyclopropyl-1-(1-ethylpyrazol-4-yl)-7-fluoranyl-indol-3-yl]sulfanyl-2-fluoranyl-benzoic acid, CALCIUM ION, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2016-07-20 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Selective Inhibition of Autotaxin Is Efficacious in Mouse Models of Liver Fibrosis.
J. Pharmacol. Exp. Ther., 360, 2017
|
|
5L01
 
 | | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | | Descriptor: | (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6MO5
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50228 complex | | Descriptor: | MAGNESIUM ION, N-[(2S)-1-(hydroxyamino)-3-methyl-3-{[(oxetan-3-yl)methyl]sulfonyl}-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOO
 
 | | Co-Crystal structure of P. aeruginosa LpxC-achn975 complex | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MO4
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOD
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50432 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-[(1S)-2-(hydroxyamino)-1-(3-methoxy-1,1-dioxo-1lambda~6~-thietan-3-yl)-2-oxoethyl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, ... | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
5TPG
 
 | | Optimization of spirocyclic proline tryptophanhydroxylase-1 inhibitors | | Descriptor: | (3S)-8-(2-amino-6-{(1R)-1-[5-chloro-3'-(methylsulfonyl)[1,1'-biphenyl]-2-yl]-2,2,2-trifluoroethoxy}pyrimidin-4-yl)-2,8-diazaspiro[4.5]decane-3-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETONITRILE, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3KDR
 
 | | The Crystal Structure of a HK97 Family Phage Portal Protein from Corynebacterium diphtheriae to 2.9A | | Descriptor: | GLYCEROL, HK97 Family Phage Portal Protein, PHOSPHATE ION, ... | | Authors: | Nocek, B, Stein, A.J, Mulligan, R, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-29 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of a HK97 Family Phage Portal Protein from Corynebacterium diphtheriae to 2.9A
To be Published
|
|
6DNA
 
 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6DNB
 
 | | Crystal structure of T110A:S256A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, GLYCEROL, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
2I91
 
 | | 60kDa Ro autoantigen in complex with a fragment of misfolded RNA | | Descriptor: | 5'-R(*C*GP*GP*UP*AP*GP*GP*CP*UP*UP*UP*UP*CP*AP*A)-3', 5'-R(*GP*CP*CP*UP*AP*CP*CP*C)-3', 60 kDa SS-A/Ro ribonucleoprotein, ... | | Authors: | Reinisch, K.M, Stein, A.J. | | Deposit date: | 2006-09-04 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4J3I
 
 | | X-ray crystal structure of bromodomain complex to 1.24 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
4J1P
 
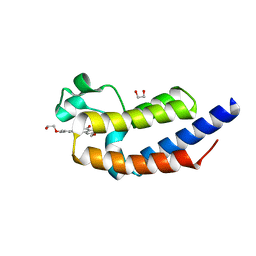 | | X-ray crystal structure of bromodomain 2 of human brd2 in complex with rvx208 to 1.08 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain containing 2, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-01 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
3LFT
 
 | |
5KPM
 
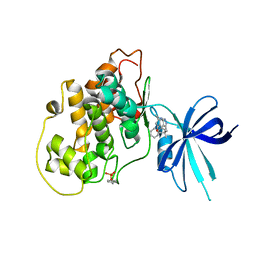 | | Glycogen Synthase Kinase 3 beta Complexed with BRD3731 | | Descriptor: | (4~{S})-3-(2,2-dimethylpropyl)-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPL
 
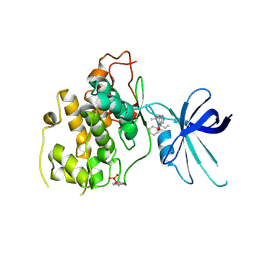 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPK
 
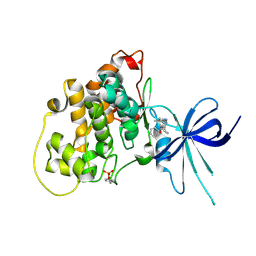 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
4MPU
 
 | | Human beta-tryptase co-crystal structure with (6S,8R)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-8-hydroxy-6-(1-hydroxycyclobutyl)-5,7-dioxaspiro[3.4]octane-6,8-dicarboxamide | | Descriptor: | (6S,8R)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-8-hydroxy-6-(1-hydroxycyclobutyl)-5,7-dioxaspiro[3.4]octane-6,8-dicarboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | White, A, Stein, A.J, Suto, R.K. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Target-Directed Self-Assembly of Homodimeric Drugs Against beta-Tryptase.
Acs Med.Chem.Lett., 9, 2018
|
|
4MPV
 
 | | Human beta-tryptase co-crystal structure with (2R,4S)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-4-hydroxy-2-(2-hydroxypropan-2-yl)-5,5-dimethyl-1,3-dioxolane-2,4-dicarboxamide | | Descriptor: | (2R,4S)-N,N'-bis[3-({4-[3-(aminomethyl)phenyl]piperidin-1-yl}carbonyl)phenyl]-4-hydroxy-2-(2-hydroxypropan-2-yl)-5,5-dimethyl-1,3-dioxolane-2,4-dicarboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Target-Directed Self-Assembly of Homodimeric Drugs Against beta-Tryptase.
Acs Med.Chem.Lett., 9, 2018
|
|
3LM9
 
 | | Crystal structure of fructokinase with ADP and Fructose bound in the active site | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ZINC ION, ... | | Authors: | Nocek, B, Stein, A, Cuff, M, Volkart, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
