4BZF
 
 | | Crystal structure of galactose mutarotase GalM from Bacillus subtilis with trehalose | | Descriptor: | ACETATE ION, ALDOSE 1-EPIMERASE, CITRIC ACID, ... | | Authors: | Vanden Broeck, A, Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Galactose Mutarotase Galm from Bacillus Subtilis with Trehalose
To be Published
|
|
4A5R
 
 | | Crystal structure of class A beta-lactamase from Bacillus licheniformis BS3 with tazobactam | | Descriptor: | BETA-LACTAMASE, CARBON DIOXIDE, CITRIC ACID, ... | | Authors: | Power, P, Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Class a Beta-Lactamase from Bacillus Licheniformis Inhibited by Tazobactam
To be Published
|
|
4D2O
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase PER- 2 | | Descriptor: | PER-2 BETA-LACTAMASE | | Authors: | Power, P, Herman, R, Ruggiero, M, Kerff, F, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2014-05-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Extended-Spectrum Beta-Lactamase Per- 2 and Insights Into the Role of Specific Residues in the Interaction with Beta-Lactams and Beta-Lactamase Inhibitors.
Antimicrob.Agents Chemother., 58, 2014
|
|
4BJQ
 
 | | Crystal structure of E. coli penicillin binding protein 3, domain V88- S165 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENICILLIN BINDING PROTEIN TRANSPEPTIDASE DOMAIN PROTEIN, SULFATE ION | | Authors: | Sauvage, E, Joris, M, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-04-19 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 3 (Pbp3) from Escherichia Coli.
Plos One, 9, 2014
|
|
4BEN
 
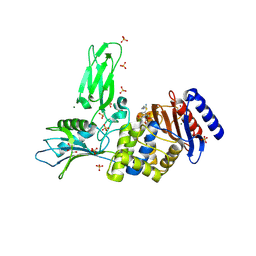 | | R39-imipenem Acyl-enzyme crystal structure | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, ... | | Authors: | Van Elder, D, Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-03-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of R39-Imipenem Acyl-Enzyme.
To be Published
|
|
3D30
 
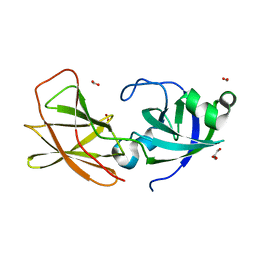 | | Structure of an expansin like protein from Bacillus Subtilis at 1.9A resolution | | Descriptor: | Expansin like protein, FORMIC ACID, GLYCEROL | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3D2Z
 
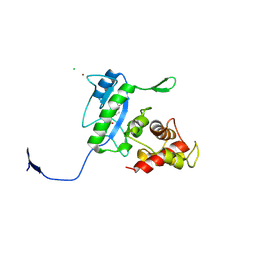 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the product L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | CHLORIDE ION, L-Ala-D-gamma-Glu-L-Lys peptide, N-acetylmuramoyl-L-alanine amidase amiD, ... | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3D2Y
 
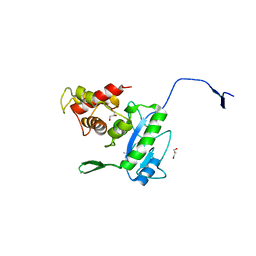 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
2WK0
 
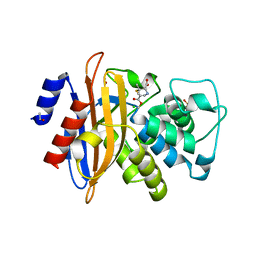 | | Crystal structure of the class A beta-lactamase BS3 inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Sauvage, E, Zervosen, A, Dive, G, Herman, R, Kerff, F, Amoroso, A, Fonze, E, Pratt, R.F, Luxen, A, Charlier, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
2WUQ
 
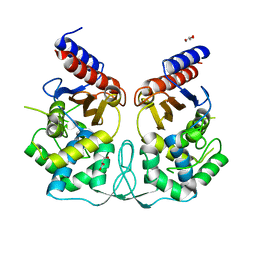 | | Crystal structure of BlaB protein from Streptomyces cacaoi | | Descriptor: | BETA-LACTAMASE REGULATORY PROTEIN BLAB, GLYCEROL | | Authors: | Dandois, S, Herman, R, Sauvage, E, Charlier, P, Joris, B, Kerff, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Blab Protein from Streptomyces Cacaoi
To be Published
|
|
2WKE
 
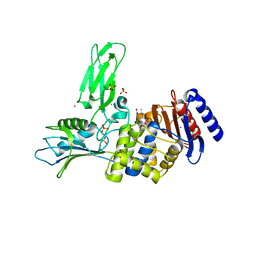 | | Crystal structure of the Actinomadura R39 DD-peptidase inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
2WKX
 
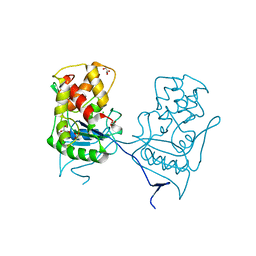 | | Crystal structure of the native E. coli zinc amidase AmiD | | Descriptor: | CHLORIDE ION, GLYCEROL, N-ACETYLMURAMOYL-L-ALANINE AMIDASE AMID, ... | | Authors: | Petrella, S, Kerff, F, Herman, R, Genereux, C, Pennartz, A, Sauvage, E, Joris, B, Charlier, P. | | Deposit date: | 2009-06-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
2AGN
 
 | | Fitting of hepatitis C virus internal ribosome entry site domains into the 15 A Cryo-EM map of a HCV IRES-80S ribosome (H. sapiens) complex | | Descriptor: | 6 nt A-RNA helix, HCV IRES DOMAIN II, HCV IRES IIIABC, ... | | Authors: | Boehringer, D, Thermann, R, Ostareck-Lederer, A, Lewis, J.D, Stark, H. | | Deposit date: | 2005-07-27 | | Release date: | 2006-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES
Structure, 13, 2005
|
|
8A0D
 
 | | Crystal structure of the major guinea pig allergen Cav p 1.0101 part of the lipocalin family | | Descriptor: | Allergen lipocalin Cav p 1 isoform 1 | | Authors: | Herman, R, Charlier, P, Janssen-Weets, B, Hilger, C, Swiontek, K. | | Deposit date: | 2022-05-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.685 Å) | | Cite: | Mammalian derived lipocalin and secretoglobin respiratory allergens strongly bind ligands with potentially immune modulating properties.
Front Allergy, 3, 2022
|
|
6YN0
 
 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | Descriptor: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
6EXS
 
 | |
7AG0
 
 | | Complex between the bone morphogenetic protein 2 and its antagonist Noggin | | Descriptor: | Bone morphogenetic protein 2, GLYCEROL, Noggin | | Authors: | Robert, C, Bruck, F, Herman, R, Vandevenne, M, Filee, P, Kerff, F, Matagne, A. | | Deposit date: | 2020-09-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural analysis of the interaction between human cytokine BMP-2 and the antagonist Noggin reveals molecular details of cell chondrogenesis inhibition.
J.Biol.Chem., 299, 2023
|
|
1W7G
 
 | | Alpha-thrombin complex with sulfated hirudin (residues 54-65) and L- Arginine template inhibitor CS107 | | Descriptor: | HIRUDIN I, N-{(1S)-1-{[4-(3-AMINOPROPYL)PIPERAZIN-1-YL]CARBONYL}-4-[(DIAMINOMETHYLENE)AMINO]BUTYL}-3-(TRIFLUOROMETHYL)BENZENESULFONAMIDE, THROMBIN | | Authors: | Remiche, J, Sauvage, E, Herman, R, Charlier, P. | | Deposit date: | 2004-09-02 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, Synthesis and Evaluation of Graftable Thrombin Inhibitors for the Preparation of Blood-Compatible Polymer Materials.
Org.Biomol.Chem., 3, 2005
|
|
2HP6
 
 | | Crystal structure of the OXA-10 W154A mutant at pH 7.5 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HPB
 
 | | Crystal structure of the OXA-10 W154A mutant at pH 9.0 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP5
 
 | | Crystal Structure of the OXA-10 W154G mutant at pH 7.0 | | Descriptor: | Beta-lactamase PSE-2, COBALT (II) ION, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP9
 
 | | Crystal Structure of the OXA-10 W154A mutant at pH 6.0 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2X02
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 AT 1.35 A RESOLUTION | | Descriptor: | BETA-LACTAMASE OXA-10, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Sauvage, E, Herman, R, Galleni, M, Charlier, P. | | Deposit date: | 2009-12-04 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Impact of the Carboxylated Lysine on the Acylation and Deacylation Step in Class D Beta-Lactamase
To be Published
|
|
2RL3
 
 | | Crystal structure of the OXA-10 W154H mutant at pH 7 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase PSE-2, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Herman, R, Sauvage, E, Guiet, R, Charlier, P, Frere, J.-M, Galleni, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2XDM
 
 | | Crystal structure of a complex between Actinomadura R39 DD peptidase and a peptidoglycan mimetic boronate inhibitor | | Descriptor: | (D-ALPHA-AMINOPIMELYLAMINO)-D-1-ETHYLBORONIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, ... | | Authors: | Rocaboy, M, Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2010-05-04 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Complex between the Actinomadura R39 Dd-Peptidase and a Peptidoglycan- Mimetic Boronate Inhibitor: Interpretation of a Transition State Analogue in Terms of Catalytic Mechanism.
Biochemistry, 49, 2010
|
|
