7KUM
 
 | |
7KUL
 
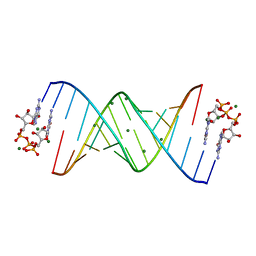 | |
7KUP
 
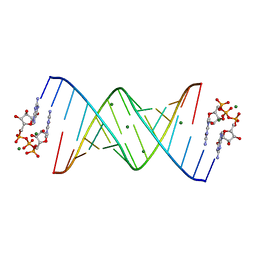 | |
7LNF
 
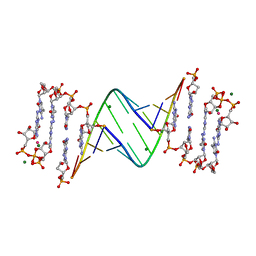 | | 3'-deoxy modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 3'-deoxy-guanosine 5'-monophosphate, 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, COBALT (II) ION, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LNG
 
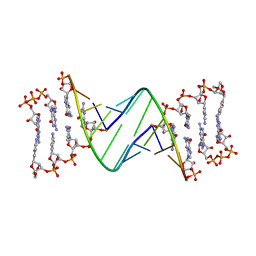 | | TNA modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, DIGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LNE
 
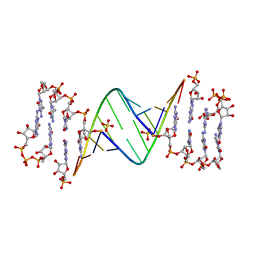 | | ANA modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | 4-AMINO-1-[(1S,3R,4R,7S)-7-HYDROXY-1-(HYDROXYMETHYL)-2,5-DIOXABICYCLO[2.2.1]HEPT-3-YL]-5-METHYLPYRIMIDIN-2(1H)-ONE, DIGUANOSINE-5'-TRIPHOSPHATE, GUANINE ARABINOSE-5'-PHOSPHATE, ... | | Authors: | Fang, Z, Giurgiu, C, Szostak, J.W. | | Deposit date: | 2021-02-07 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7U8B
 
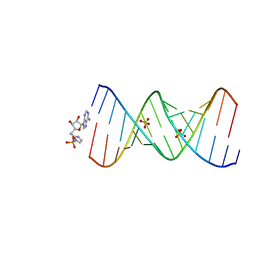 | | Product of 14mer primer with activated asymmetric GA dimer diastereomer 1 | | Descriptor: | 5'-O-[(S)-(2-amino-1H-imidazol-1-yl)(hydroxy)phosphoryl]adenosine, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3'), SULFATE ION | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Understanding the metal ion function in the RNA non-enzymatic extesion with phosphorothioates
To Be Published
|
|
1GMY
 
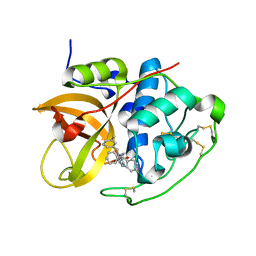 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
6X5D
 
 | |
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W4E
 
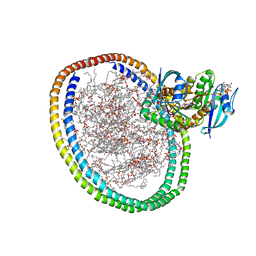 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7KU4
 
 | |
7KWK
 
 | |
3HG8
 
 | |
3IJK
 
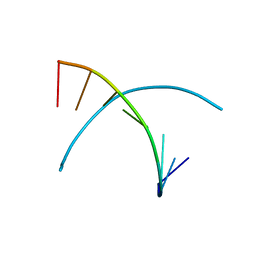 | | 5-OMe modified DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Zhang, W, Hassan, A.E.A, Gan, J, Huang, Z. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
3LTU
 
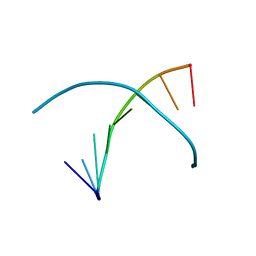 | | 5-SeMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5S)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
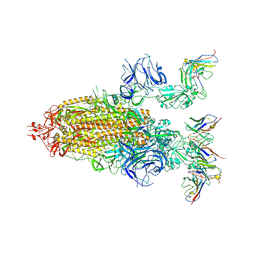
8DZI
 
 | |
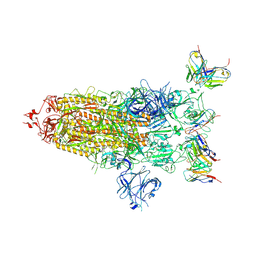
8DZH
 
 | |
9LAE
 
 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies 9G11 and 3E2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 3E2, Heavy chain of 9G11, ... | | Authors: | Jiang, Y, Sun, H, Zheng, Q, Li, S. | | Deposit date: | 2025-01-02 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural insight into broadening SARS-CoV-2 neutralization by an antibody cocktail harbouring both NTD and RBD potent antibodies.
Emerg Microbes Infect, 13, 2024
|
|
5HQA
 
 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQ4
 
 | | A Glycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQC
 
 | | A Glycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQB
 
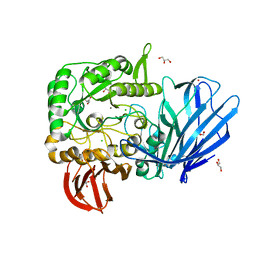 | | A Glycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
7RQT
 
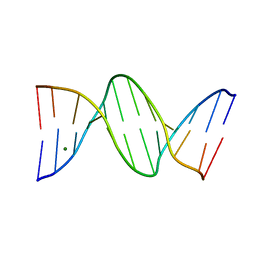 | | THE B-DNA DODECAMER With HIGH RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chen, C, Huang, Z. | | Deposit date: | 2021-08-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | 2'-beta-Selenium Atom on Thymidine to Control beta-Form DNA Conformation and Large Crystal Formation
Cryst.Growth Des., 2022
|
|
7R60
 
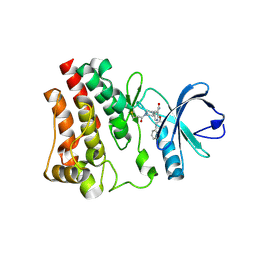 | | BTK in complex with 18A | | Descriptor: | 2-(4-phenoxyphenoxy)-5-[(3R)-1-(prop-2-enoyl)piperidin-3-yl]pyridine-3-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Covalent Bruton's Tyrosine Kinase Inhibitors with Decreased CYP2C8 Inhibitory Activity.
Chemmedchem, 16, 2021
|
|
