7M6I
 
 | |
7M6D
 
 | |
7K8R
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C135 | | Descriptor: | C135 Fab Heavy Chain, C135 Fab Light Chain, GLYCEROL | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8O
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C002 | | Descriptor: | C002 Fab Heavy Chain, C002 Fab Light Chain, GLYCEROL, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8W
 
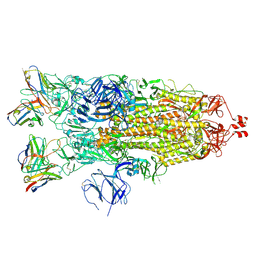 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C119 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C119 Fab Heavy Chain, ... | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8T
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C002 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | Authors: | Malyutin, A.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
8D47
 
 | |
8D48
 
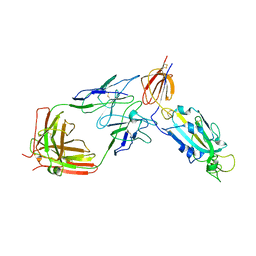 | | sd1.040 Fab in complex with SARS-CoV-2 Spike 2P glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, sd1.040 Fab heavy chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human neutralizing antibodies to cold linear epitopes and subdomain 1 of the SARS-CoV-2 spike glycoprotein.
Sci Immunol, 8, 2023
|
|
7R8O
 
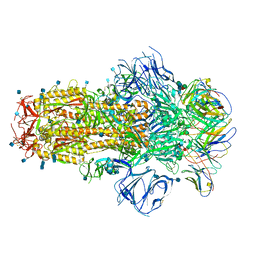 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C548 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8M
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C032 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7S0B
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | Authors: | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
6UDK
 
 | | HIV-1 bNAb 1-55 in complex with modified BG505 SOSIP-based immunogen RC1 and 10-1074 | | Descriptor: | 1-55 Fab Heavy Chain, 1-55 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UDJ
 
 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
9BJ4
 
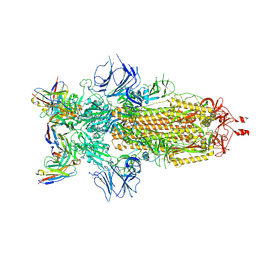 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C952 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C952 Heavy Chain, C952 Light Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
9BJ2
 
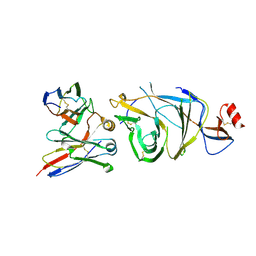 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1533 (local refinement of NTD and C1533) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1533 Heavy Chain, C1533 Light Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
9BJ3
 
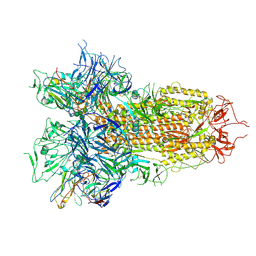 | | Structure of the SARS-CoV-2 S 6P trimer complex with the human neutralizing antibody Fab fragment, C1596 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1596 Fab Heavy Chain, ... | | Authors: | Rubio, A.A, Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Bispecific antibodies targeting the N-terminal and receptor binding domains potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 17, 2025
|
|
7RKS
 
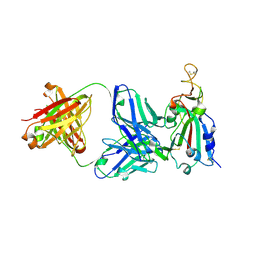 | | Structure of the SARS-CoV receptor binding domain in complex with the human neutralizing antibody Fab fragment, C118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Antibody Fab Heavy Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKU
 
 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7N3E
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C032 | | Descriptor: | C032 Fab Heavy Chain, C032 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3G
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C098 | | Descriptor: | C098 Fab Heavy Chain, C098 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3H
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C099 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3F
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C080 | | Descriptor: | C080 Fab Heavy Chain, C080 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3I
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment C098 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C098 Fab heavy chain, C098 Fab light chain, ... | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7UGM
 
 | |
5JK7
 
 | | The X-ray structure of the DDB1-DCAF1-Vpr-UNG2 complex | | Descriptor: | DNA damage-binding protein 1, Protein VPRBP, Protein Vpr, ... | | Authors: | Calero, G, Ahn, J, Wu, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction.
Nat.Struct.Mol.Biol., 23, 2016
|
|
