7N1F
 
 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 binds to YLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1D
 
 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 | | Descriptor: | pYLQ7 T cell receptor alpha chain, pYLQ7 T cell receptor beta chain | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
3NM1
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
7N1E
 
 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 binds to RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1C
 
 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 | | Descriptor: | pRLQ3 T cell receptor alpha chain, pRLQ3 T cell receptor beta chain | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
5F6D
 
 | | Crystal structure of Ubc9 (K48A/K49A/E54A) complexed with Fragment 6 | | Descriptor: | 6~{H}-benzo[c][1,2]benzothiazine 5,5-dioxide, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-05 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3NGK
 
 | |
5EXA
 
 | | Small-molecule stabilization of the 14-3-3/Gab2 PPI interface | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Fusicoccin A-THF derivative, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-04 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface.
Chemmedchem, 11, 2016
|
|
5F6I
 
 | |
5F24
 
 | | Crystal structure of dual specific IMPase/NADP phosphatase bound with D-inositol-1-phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-MYO-INOSITOL-1-PHOSPHATE, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1BEV
 
 | | BOVINE ENTEROVIRUS VG-5-27 | | Descriptor: | BOVINE ENTEROVIRUS COAT PROTEINS VP1 TO VP4, MYRISTIC ACID, SULFATE ION | | Authors: | Smyth, M, Tate, J, Lyons, C, Hoey, E, Martin, S, Stuart, D. | | Deposit date: | 1996-04-03 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Implications for viral uncoating from the structure of bovine enterovirus.
Nat.Struct.Biol., 2, 1995
|
|
5F15
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5F5F
 
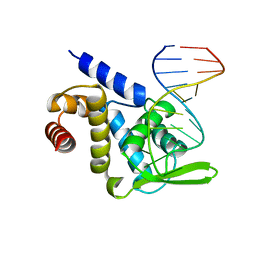 | | X-ray structure of Roquin ROQ domain in complex with a Selex-derived hexa-loop RNA motif | | Descriptor: | RNA (5'-R(P*UP*GP*AP*CP*UP*GP*CP*GP*UP*UP*UP*UP*AP*GP*GP*AP*GP*UP*UP*A)-3'), Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2015-12-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roquin recognizes a non-canonical hexaloop structure in the 3'-UTR of Ox40.
Nat Commun, 7, 2016
|
|
5F6W
 
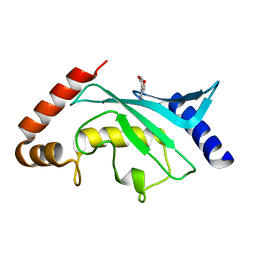 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol) | | Descriptor: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5EZZ
 
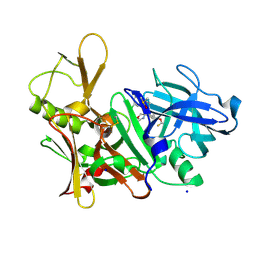 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH (4S)-4-[3-(5-chloro-3-pyridyl)phenyl]-4-[4-(difluoromethoxy)-3-methyl-phenyl]-5H-oxazol-2-amine | | Descriptor: | (4~{S})-4-[4-[bis(fluoranyl)methoxy]-3-methyl-phenyl]-4-[3-(5-chloranylpyridin-3-yl)phenyl]-5~{H}-1,3-oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EW8
 
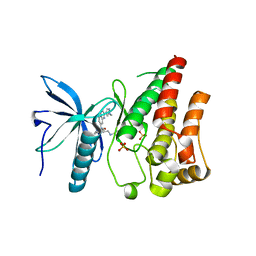 | | FIBROBLAST GROWTH FACTOR RECEPTOR 1 IN COMPLEX WITH JNJ-4275693 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}'-(3,5-dimethoxyphenyl)-~{N}'-[3-(1-methylpyrazol-4-yl)quinoxalin-6-yl]-~{N}-propan-2-yl-ethane-1,2-diamine | | Authors: | Ogg, D, Breed, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use.
Oncotarget, 7, 2016
|
|
5EWZ
 
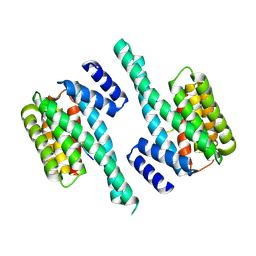 | | Small-molecule stabilization of the 14-3-3/Gab2 PPI interface | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, GRB2-associated-binding protein 2 | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-04 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface.
Chemmedchem, 11, 2016
|
|
5EYH
 
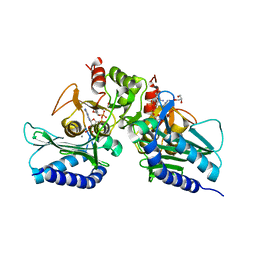 | | Crystal Structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ at pH 7.0 | | Descriptor: | CALCIUM ION, GLYCEROL, Inositol monophosphatase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3NEQ
 
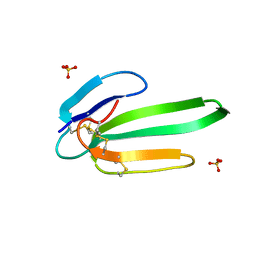 | | Crystal structure of the chimeric muscarinic toxin MT7 with loop 3 from MT1 | | Descriptor: | SULFATE ION, Three-finger muscarinic toxin 7 | | Authors: | Stura, E.A, Servent, D, Menez, R, Mournier, G, Menez, A, Fruchart-Gaillard, C. | | Deposit date: | 2010-06-09 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors.
Plos One, 7, 2012
|
|
3NL5
 
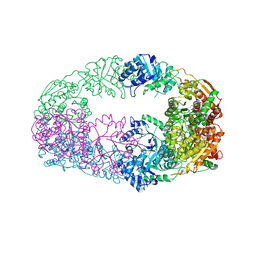 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
1BFD
 
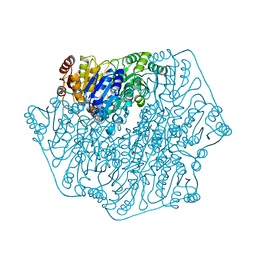 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hasson, M.S, Muscate, A, Mcleish, M.J, Polovnikova, L.S, Gerlt, J.A, Kenyon, G.L, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-04-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
Biochemistry, 37, 1998
|
|
5F03
 
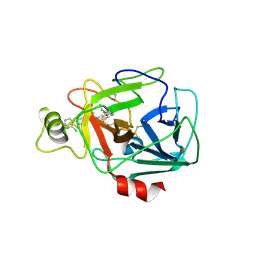 | | TRYPTASE B2 IN COMPLEX WITH 5-(3-Aminomethyl-phenoxymethyl)-3-[3-(2-chloro-pyridin-3-ylethynyl)-phenyl]-oxazolidin-2-one; compound with trifluoro-acetic acid | | Descriptor: | (5~{S})-5-[[3-(aminomethyl)phenoxy]methyl]-3-[3-[2-(2-chloranylpyridin-3-yl)ethynyl]phenyl]-1,3-oxazolidin-2-one, Tryptase beta-2 | | Authors: | Banner, D, Benz, J, Joseph, C, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5F6V
 
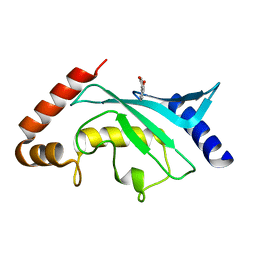 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol from fragment cocktail screen) | | Descriptor: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5EXZ
 
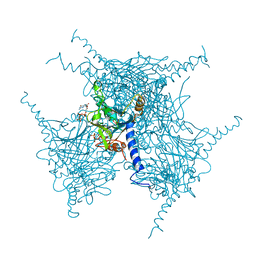 | | Crystal structure of purified recombinant CPV1 Polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Boudes, M, Garriga, D, Coulibaly, F. | | Deposit date: | 2015-11-24 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A pipeline for structure determination of in vivo-grown crystals using in cellulo diffraction.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYG
 
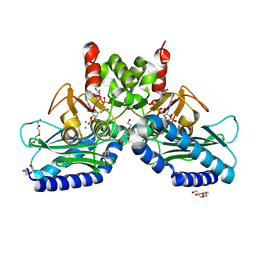 | | Crystal structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
