4O9B
 
 | | The Structure of CC1-IH in human STIM1. | | Descriptor: | CADMIUM ION, Stromal interaction molecule 1 | | Authors: | Cui, B, Yang, X, Li, S, Shen, Y. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1.
Plos One, 8, 2013
|
|
8Q6T
 
 | |
7WHB
 
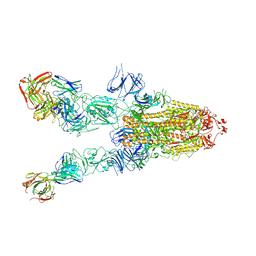 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Ge, J.W, Wang, X.Q. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
4JII
 
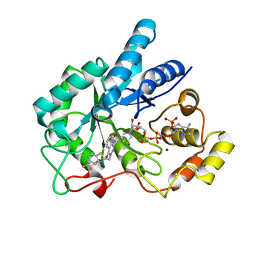 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
8GJY
 
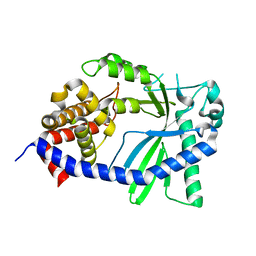 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJW
 
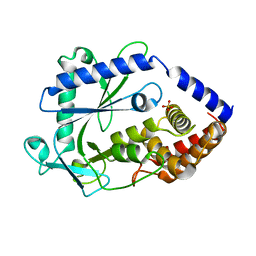 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJX
 
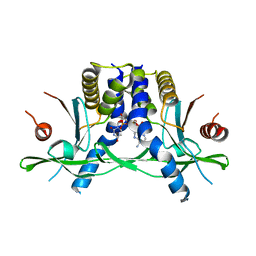 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJZ
 
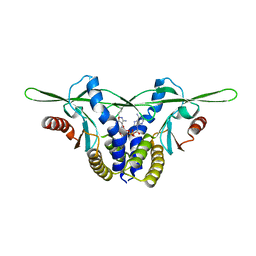 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7YHD
 
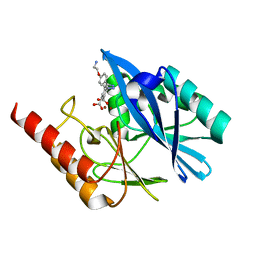 | | Crystal structure of VIM-2 MBL in complex with 3-(4-(4-(2-aminoethoxy)phenyl)-1H-1,2,3-triazol-1-yl)phthalic acid | | Descriptor: | 3-[4-[4-(2-azanylethoxy)phenyl]-1,2,3-triazol-1-yl]phthalic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YHB
 
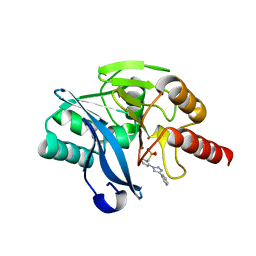 | | Crystal structure of VIM-2 MBL in complex with (2-(4-phenyl-1H-1,2,3-triazol-1-yl)benzyl)phosphonic acid | | Descriptor: | Beta-lactamase class B VIM-2, ZINC ION, [2-(4-phenyl-1,2,3-triazol-1-yl)phenyl]methylphosphonic acid | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YH9
 
 | | Crystal structure of IMP-1 MBL in complex with 3-(4-benzyl-1H-1,2,3-triazol-1-yl)phthalic acid | | Descriptor: | 3-[4-(phenylmethyl)-1,2,3-triazol-1-yl]phthalic acid, Beta-lactamase class B IMP-1, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YHC
 
 | | Crystal structure of VIM-2 MBL in complex with 3-(4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl)phthalic acid | | Descriptor: | 3-[4-(3-aminophenyl)-1,2,3-triazol-1-yl]phthalic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
7YHA
 
 | | Crystal structure of IMP-1 MBL in complex with (3-(4-(p-tolyl)-1H-1,2,3-triazol-1-yl)benzyl)phosphonic acid | | Descriptor: | Beta-lactamase class B IMP-1, ZINC ION, [3-[4-(4-methylphenyl)-1,2,3-triazol-1-yl]phenyl]methylphosphonic acid | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
5H5Z
 
 | | Crystal structure of bony fish MHC class I, peptide and B2m II | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide chain | | Authors: | Chen, Z, Zhang, N, Qi, J, Li, X, Chen, R, Wang, Z, Gao, F.G, Xia, C. | | Deposit date: | 2016-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Mechanism of beta 2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish.
Iscience, 23, 2020
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NHL
 
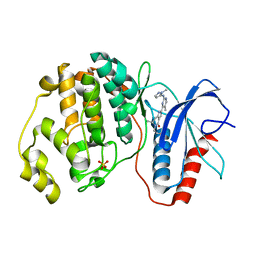 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{R})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
6J8R
 
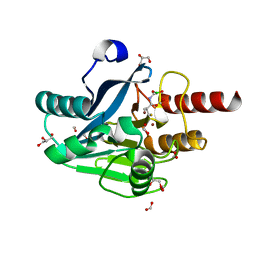 | | Metallo-Beta-Lactamase VIM-2 in complex with Dual MBL/SBL Inhibitor MS01 | | Descriptor: | Beta-lactamase class B VIM-2, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6J8Q
 
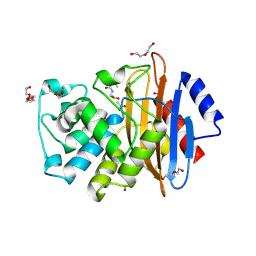 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor WL-001 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
8HTR
 
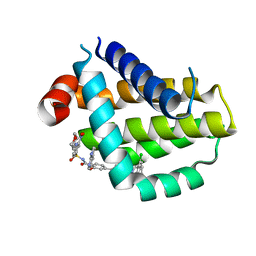 | | Crystal structure of Bcl2 in complex with S-9c | | Descriptor: | 4-[4-[(2~{S})-2-(2-chlorophenyl)pyrrolidin-1-yl]phenyl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
8HTS
 
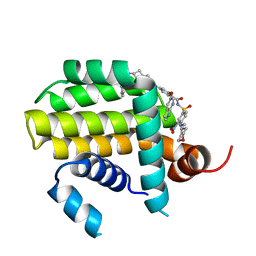 | | Crystal structure of Bcl2 in complex with S-10r | | Descriptor: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
5J1E
 
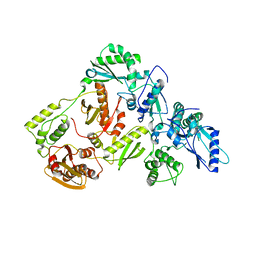 | | Crystal Structure of a Hydroxypyridone Carboxylic Acid Active-Site RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 5-hydroxy-4-oxo-1-[(4'-sulfamoyl[1,1'-biphenyl]-4-yl)methyl]-1,4-dihydropyridine-3-carboxylic acid, HIV-1 REVERSE TRANSCRIPTASE P51 DOMAIN, HIV-1 REVERSE TRANSCRIPTASE P66 DOMAIN, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H.
J.Med.Chem., 59, 2016
|
|
5YKI
 
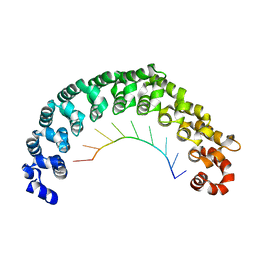 | | Crystal structure of the engineered nine-repeat PUF domain in complex with cognate 9nt-RNA | | Descriptor: | Pumilio homolog 1, RNA (5'-R(*UP*GP*UP*UP*GP*UP*AP*UP*A)-3') | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
5YKH
 
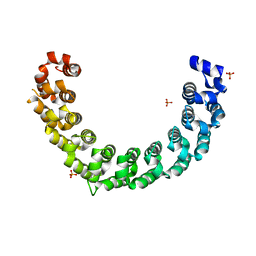 | | Crystal structure of the engineered nine-repeat PUF domain | | Descriptor: | PHOSPHATE ION, Pumilio homolog 1 | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
6JN4
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor WL-001 | | Descriptor: | ACETATE ION, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6JN5
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor MS23 | | Descriptor: | Serine Beta-Lactamase KPC-2, [(S)-(4-fluorophenyl)-[[(2S)-2-methyl-3-sulfanyl-propanoyl]amino]methyl]boronic acid | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
