3G9K
 
 | | Crystal structure of Bacillus anthracis transpeptidase enzyme CapD | | Descriptor: | Capsule biosynthesis protein capD, GLUTAMIC ACID | | Authors: | Zhang, R, Wu, R, Richter, S, Anderson, V.J, Missiakas, D, Joachimiak, A. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Bacillus anthracis Transpeptidase Enzyme CapD.
J.Biol.Chem., 284, 2009
|
|
1ILV
 
 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
3SWJ
 
 | | Crystal structure of Campylobacter jejuni ChuZ | | Descriptor: | AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein | | Authors: | Hu, Y. | | Deposit date: | 2011-07-14 | | Release date: | 2011-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
8D6K
 
 | | Sco GlgEI-V279S in complex with cyclohexyl carbasugar | | Descriptor: | (1R,4S,5S,6R)-4-(cyclohexylamino)-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER | | Authors: | Jayasinghe, T.J, Ronning, D.R. | | Deposit date: | 2022-06-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Synthesis of C 7 /C 8 -cyclitols and C 7 N-aminocyclitols from maltose and X-ray crystal structure of Streptomyces coelicolor GlgEI V279S in a complex with an amylostatin GXG-like derivative.
Front Chem, 10, 2022
|
|
7CBJ
 
 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | Descriptor: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
1L3L
 
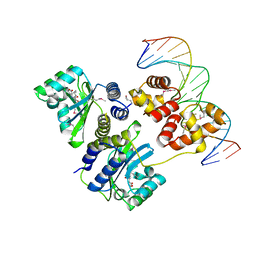 | | Crystal structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*GP*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP*CP*TP*GP*CP*AP*CP*AP*TP*C)-3', Transcriptional activator protein traR | | Authors: | Zhang, R, Pappas, T, Brace, J.L, Miller, P.C, Oulmassov, T, Molyneaux, J.M, Anderson, J.C, Bashkin, J.K, Winans, S.C, Joachimiak, A. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA.
Nature, 417, 2002
|
|
1P99
 
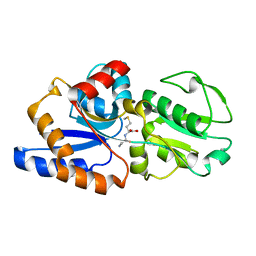 | | 1.7A crystal structure of protein PG110 from Staphylococcus aureus | | Descriptor: | GLYCINE, Hypothetical protein PG110, METHIONINE | | Authors: | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions.
Biochemistry, 43, 2004
|
|
1YWQ
 
 | |
7EAU
 
 | | SIB1, an effector of Colletotrichum orbiculare | | Descriptor: | SIN1 | | Authors: | Mori, M, Ohki, S, Kurita, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Fungal effector SIB1 of Colletotrichum orbiculare has unique structural features and can suppress plant immunity in Nicotiana benthamiana.
J.Biol.Chem., 297, 2021
|
|
3NRV
 
 | |
1TMH
 
 | |
7VEG
 
 | | Understanding NH-pi interaction between Gln and Phe | | Descriptor: | peptide, ACETYLAMINO-ACETIC ACID | | Authors: | Fan, S, Xu, F. | | Deposit date: | 2021-09-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Achievability of an NH-pi Interaction between Gln and Phe in a Crystal Structure of a Collagen-like Peptide.
Biomolecules, 12, 2022
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5YPR
 
 | | Crystal Structure of PSD-95 SH3-GK domain in complex with a synthesized inhibitor | | Descriptor: | Disks large homolog 4, Synthesized GK inhibitor | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhu, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
6CZN
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Z2OHTPE and a glucocorticoid receptor-interacting protein 1 NR box II peptide | | Descriptor: | 4,4'-[(1R,2R)-1-phenylbutane-1,2-diyl]diphenol, Estrogen receptor, GRIP Peptide | | Authors: | Fanning, S.W, Han, R, Maximov, P, Jordan, V.C, Greene, G.L. | | Deposit date: | 2018-04-09 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Z2OHTPE and a glucocorticoid receptor-interacting protein 1 NR box II peptide
To Be Published
|
|
7XS8
 
 | |
7XSA
 
 | |
7XSB
 
 | |
7XSC
 
 | |
5YBE
 
 | | Structure of KANK1/KIF21A complex | | Descriptor: | 1,2-ETHANEDIOL, KIF21A, Kank1 protein | | Authors: | Weng, Z.F, Shang, Y, Yao, D.Q, Zhu, J.W, Zhang, R.G. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Structural analyses of key features in the KANK1/KIF21A complex yield mechanistic insights into the cross-talk between microtubules and the cell cortex.
J. Biol. Chem., 293, 2018
|
|
7XMN
 
 | | Structure of SARS-CoV-2 ORF8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein, ... | | Authors: | Chen, X, Xu, W. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylated, Lipid-Binding, CDR-Like Domains of SARS-CoV-2 ORF8 Indicate Unique Sites of Immune Regulation.
Microbiol Spectr, 11, 2023
|
|
6V8T
 
 | |
3KA0
 
 | |
6VPF
 
 | |
