4NPL
 
 | |
1NOT
 
 | | THE 1.2 ANGSTROM STRUCTURE OF G1 ALPHA CONOTOXIN | | Descriptor: | GI ALPHA CONOTOXIN | | Authors: | Guddat, L.W, Shan, L, Martin, J.L, Edmundson, A.B, Gray, W.R. | | Deposit date: | 1996-05-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Three-dimensional structure of the alpha-conotoxin GI at 1.2 A resolution
Biochemistry, 35, 1996
|
|
4NPM
 
 | |
1RKN
 
 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
3NK4
 
 | |
3NK3
 
 | | Crystal structure of full-length sperm receptor ZP3 at 2.6 A resolution | | Descriptor: | CITRATE ANION, Zona pellucida 3, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Monne, M, Jovine, L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into Egg Coat Assembly and Egg-Sperm Interaction from the X-Ray Structure of Full-Length ZP3.
Cell(Cambridge,Mass.), 143, 2010
|
|
7WBI
 
 | | BF2*1901-FLU | | Descriptor: | Beta-2-microglobulin, ILE-ARG-HIS-GLU-ASN-ARG-MET-VAL-LEU, MHC class I alpha chain 2 | | Authors: | Liu, W.J. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
7WBG
 
 | | BF2*1901/RY8 | | Descriptor: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I alpha chain 2 | | Authors: | Liu, W.J. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
7NZN
 
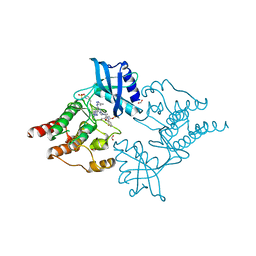 | | Structure of RET kinase domain bound to inhibitor JB-48 | | Descriptor: | 2-[4-[[4-[1-[2-(dimethylamino)ethyl]pyrazol-4-yl]-6-[(3-methyl-1~{H}-pyrazol-5-yl)amino]pyrimidin-2-yl]amino]phenyl]-~{N}-(3-fluorophenyl)ethanamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2021-03-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-Trisubstituted Pyrimidine Derivatives as Type I RET and RET Gatekeeper Mutant Inhibitors with a Novel Kinase Binding Pose.
J.Med.Chem., 65, 2022
|
|
6O3G
 
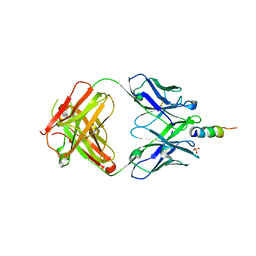 | |
6O3D
 
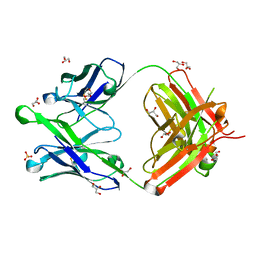 | |
6O3J
 
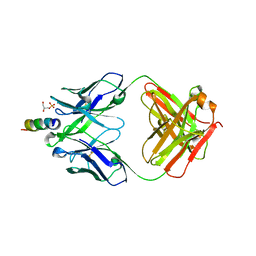 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.416 Å) | | Cite: | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
6O3L
 
 | |
6O41
 
 | |
6O42
 
 | |
6O3K
 
 | |
6O3U
 
 | |
7PH1
 
 | | Trypsin in complex with BPTI mutant (2S)-2-amino-4-monofluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Loll, B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Water Network in the Binding Pocket of Fluorinated BPTI-Trypsin Complexes─Insights from Simulation and Experiment.
J.Phys.Chem.B, 126, 2022
|
|
7NS6
 
 | |
6UKA
 
 | | Crystal structure of RHOG and ELMO complex | | Descriptor: | Engulfment and cell motility protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jo, C.H, Killoran, R.C, Smith, M.J. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6ZXN
 
 | |
7VQD
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and geranylgeranyl pyrophosphate. | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, NerylNeryl pyrophosphate, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQC
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with pyrophosphate | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, PYROPHOSPHATE 2-, SULFATE ION | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQB
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and dimethylallyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQA
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with dimethylallyl diphosphate. | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
