1A5E
 
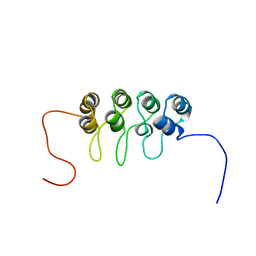 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
5SV7
 
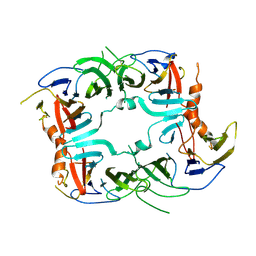 | | The Crystal structure of a chaperone | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wang, P, Li, J, Sha, B. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4G5Q
 
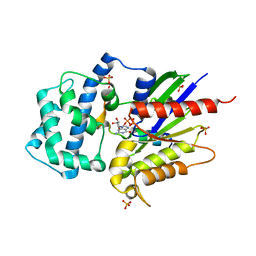 | | Structure of LGN GL4/Galphai1 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from G alpha i.
J.Biol.Chem., 287, 2012
|
|
5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | Descriptor: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | Authors: | Gorman, J, Li, J, Kwong, P.D. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
2CH2
 
 | | Structure of the Anopheles gambiae 3-hydroxykynurenine transaminase in complex with inhibitor | | Descriptor: | 3-HYDROXYKYNURENINE TRANSAMINASE, 4-(2-AMINOPHENYL)-4-OXOBUTANOIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Garavaglia, S, Giovenzana, G.B, Arca, B, Li, J, Rizzi, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-03-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Anopheles Gambiae 3-Hydroxykynurenine Transaminase
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7DL2
 
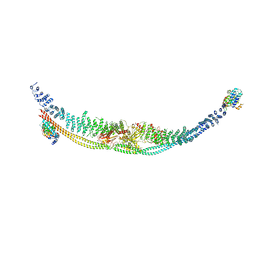 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
5U2L
 
 | | Crystal structure of the Hsp104 N-terminal domain from Candida albicans | | Descriptor: | Heat shock protein 104 | | Authors: | Wang, P, Li, J, Sha, B. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6555 Å) | | Cite: | Crystal structures of Hsp104 N-terminal domains from Saccharomyces cerevisiae and Candida albicans suggest the mechanism for the function of Hsp104 in dissolving prions.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5U2U
 
 | |
2CH1
 
 | | Structure of Anopheles gambiae 3-hydroxykynurenine transaminase | | Descriptor: | 3-HYDROXYKYNURENINE TRANSAMINASE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Garavaglia, S, Giovenzana, G.B, Arca, B, Li, J, Rizzi, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Anopheles Gambiae 3-Hydroxykynurenine Transaminase
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
8ZC9
 
 | | The Cryo-EM structure of DSR2-Tail tube-NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein, tail tube protein | | Authors: | Wang, R, Xu, Q, Wu, Z, Li, J, Yang, R, Shi, Z, Li, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
1OLS
 
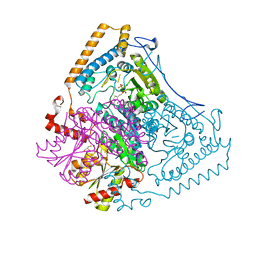 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
3RHZ
 
 | |
2Q8I
 
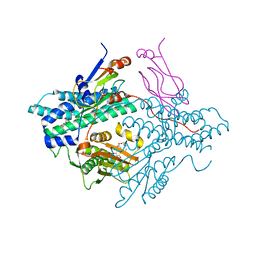 | | Pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug radicicol | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, GLYCEROL, ... | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
8U8H
 
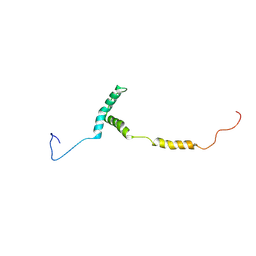 | |
8U95
 
 | | The structure of myosin heavy chain from Drosophila melanogaster flight muscle thick filaments | | Descriptor: | Myosin heavy chain, isoform U | | Authors: | Abbasi Yeganeh, F, Rastegarpouyani, H, Li, J, Taylor, K.A. | | Deposit date: | 2023-09-18 | | Release date: | 2023-10-18 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 angstrom Resolution Reveals New Details of Non-Myosin Proteins.
Int J Mol Sci, 24, 2023
|
|
2Q8H
 
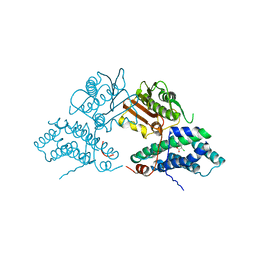 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with dichloroacetate (DCA) | | Descriptor: | DICHLORO-ACETIC ACID, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
1B34
 
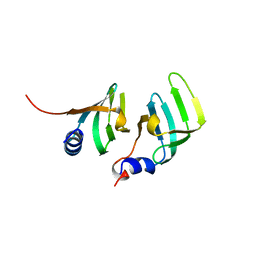 | | CRYSTAL STRUCTURE OF THE D1D2 SUB-COMPLEX FROM THE HUMAN SNRNP CORE DOMAIN | | Descriptor: | PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D1), PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D2) | | Authors: | Walke, S, Young, R.J, Kambach, C, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-17 | | Release date: | 2000-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
2Q8G
 
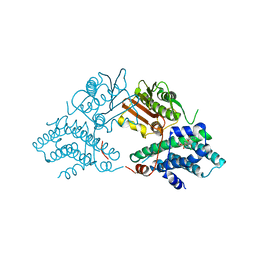 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with glucose-lowering drug AZD7545 | | Descriptor: | 4-[(3-CHLORO-4-{[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]AMINO}PHENYL)SULFONYL]-N,N-DIMETHYLBENZAMIDE, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2Q8F
 
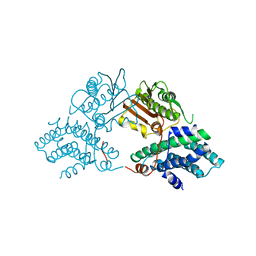 | | Structure of pyruvate dehydrogenase kinase isoform 1 | | Descriptor: | POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
2QLR
 
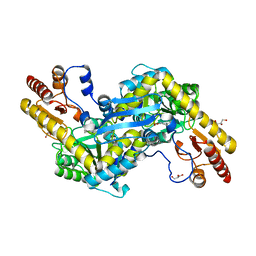 | |
2A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
2R2N
 
 | | The crystal structure of human kynurenine aminotransferase II in complex with kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2007-08-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase II.
J.Biol.Chem., 283, 2008
|
|
2QTW
 
 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8WSJ
 
 | |
