9J7U
 
 | | H176A mutant of human G6PC1 in complex with G6P | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-O-phosphono-beta-D-glucopyranose, Glucose-6-phosphatase catalytic subunit 1 | | Authors: | Jiang, D.H, Xia, Z.Y. | | Deposit date: | 2024-08-19 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into glucose-6-phosphate recognition and hydrolysis by human G6PC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8U1D
 
 | | Cryo-EM structure of vaccine-elicited CD4 binding site antibody DH1285 bound to HIV-1 CH505TFchim.6R.SOSIP.664v4.1 Env Local Refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1285 Heavy Chain, DH1285 Light Chain, ... | | Authors: | Thakur, B, Stalls, V.D, Acharya, P. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Vaccine induction of CD4-mimicking HIV-1 broadly neutralizing antibody precursors in macaques.
Cell, 187, 2024
|
|
8XXS
 
 | | Crystal structure of PDE4D catalytic domain complexed with L11 | | Descriptor: | 2-[4-[bis(fluoranyl)methoxy]-3-(cyclopropylmethoxy)phenyl]-1-benzofuran-6-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wu, D, Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.10000944 Å) | | Cite: | 5-hydroxymethylcytosine features of portal venous blood predict metachronous liver metastases of colorectal cancer and reveal phosphodiesterase 4 as a therapeutic target.
Clin Transl Med, 15, 2025
|
|
6JJB
 
 | | BRD4 in complex with ZZM1 | | Descriptor: | 2-methoxy-N-(1-methyl-2-oxidanylidene-benzo[cd]indol-6-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
6JJ3
 
 | | BRD4 in complex with 138A | | Descriptor: | 2-methoxy-N-[2-methyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
8YX3
 
 | |
8YX4
 
 | |
8YX2
 
 | |
8YX5
 
 | | Crystal Structure of SARS CoV-2 Papain-like Protease PLpro-C111S in Complex with GZNL-P35 | | Descriptor: | 5-[(1~{R},5~{S})-3,6-diazabicyclo[3.1.1]heptan-3-yl]-2-methyl-~{N}-[1-(1-methyl-2-oxidanylidene-benzo[cd]indol-6-yl)cyclopropyl]benzamide, GLYCEROL, Papain-like protease nsp3, ... | | Authors: | Lu, Y, Shang, J. | | Deposit date: | 2024-04-02 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of orally bioavailable SARS-CoV-2 papain-like protease inhibitor as a potential treatment for COVID-19.
Nat Commun, 15, 2024
|
|
5T0K
 
 | | Structure of G9a SET-domain with H3K9M mutant peptide and SAM | | Descriptor: | H3K9 mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading.
Elife, 5, 2016
|
|
5T0M
 
 | |
5UD8
 
 | | Crystal Structure of Mutant Ig-like Domain | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
5UD7
 
 | | Crystal Structure of Wild-Type Ig-like Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, SULFATE ION, ... | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20002246 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
6CZS
 
 | |
6CZK
 
 | | Crystal structure of wild-type human pro-cathepsin H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pro-cathepsin H, ... | | Authors: | Huang, X, Hao, Y. | | Deposit date: | 2018-04-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structures of human procathepsin H.
PLoS ONE, 13, 2018
|
|
2NSQ
 
 | | Crystal structure of the C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase NEDD4-like protein, GLYCEROL | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein
To be Published
|
|
6AKY
 
 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 34 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-3-yl)propyl]cyclohexane-1-carboxamide, C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
6K1M
 
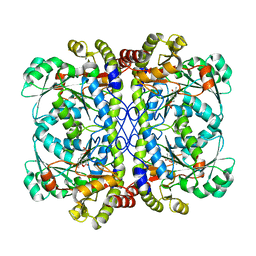 | | Engineered form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
6AKX
 
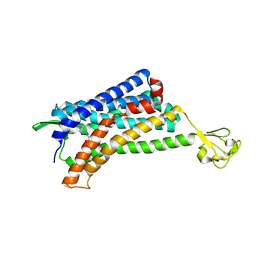 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 21 | | Descriptor: | C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-2-yl)propyl]cyclopentanecarboxamide, NITRATE ION, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
6K1L
 
 | | E53A mutant of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, PYRUVIC ACID | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
6K1O
 
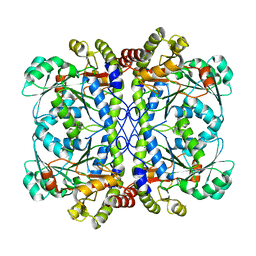 | | Apo form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
8J4U
 
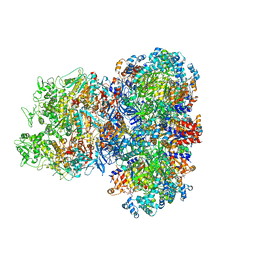 | | Structure of HerA-Sir2 complex from Escherichia coli Nezha system | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate hydrolase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Q, Yu, Y. | | Deposit date: | 2023-04-20 | | Release date: | 2024-01-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Multiple enzymatic activities of a Sir2-HerA system cooperate for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8KAG
 
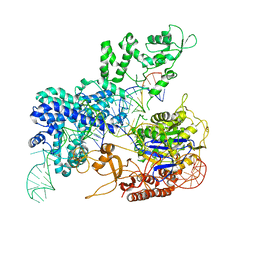 | | Crystal structure of SpyCas9 in complex with sgRNA and target RNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, RNA (5'-R(*CP*CP*AP*CP*UP*UP*CP*AP*AP*UP*UP*AP*GP*AP*AP*CP*AP*CP*GP*GP*AP*CP*C)-3'), RNA (98-MER) | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 43, 2025
|
|
8KAK
 
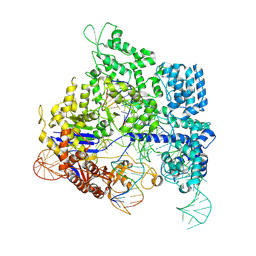 | | Crystal structure of SpyCas9 in complex with sgRNA and 18nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (26-MER), DNA (5'-D(*TP*TP*TP*AP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 43, 2025
|
|
