8FH4
 
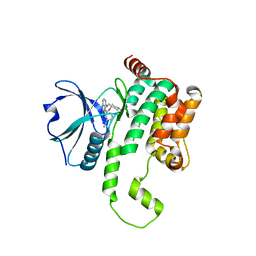 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
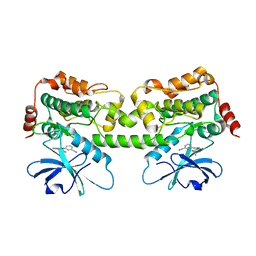 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FJZ
 
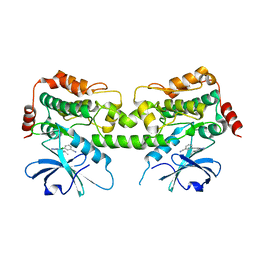 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
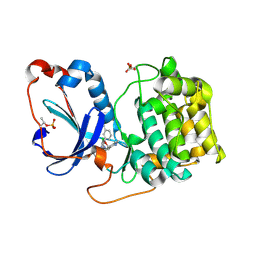 | | PKCeta kinase domain in complex with compound 2 | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
7XRF
 
 | | Crystal structaure of DgpB/C complex | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB, MANGANESE (II) ION | | Authors: | Ma, M, He, P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural mechanism of a dual-functional enzyme DgpA/B/C as both a C -glycoside cleaving enzyme and an O - to C -glycoside isomerase.
Acta Pharm Sin B, 13, 2023
|
|
7XRE
 
 | | Crystal structure of DgpA | | Descriptor: | DgpA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ma, W, He, P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural mechanism of a dual-functional enzyme DgpA/B/C as both a C -glycoside cleaving enzyme and an O - to C -glycoside isomerase.
Acta Pharm Sin B, 13, 2023
|
|
7XR9
 
 | | Crystal structure of DgpA with glucose | | Descriptor: | DgpA, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, beta-D-glucopyranose | | Authors: | Ma, W, He, P. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural mechanism of a dual-functional enzyme DgpA/B/C as both a C -glycoside cleaving enzyme and an O - to C -glycoside isomerase.
Acta Pharm Sin B, 13, 2023
|
|
5UD8
 
 | | Crystal Structure of Mutant Ig-like Domain | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
6MQC
 
 | |
5UD7
 
 | | Crystal Structure of Wild-Type Ig-like Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, SULFATE ION, ... | | Authors: | Sudom, A, Min, X, Wang, Z. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.20002246 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
6MQM
 
 | |
6MQR
 
 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6N16
 
 | |
8ERC
 
 | |
6MQE
 
 | |
6MQS
 
 | |
6U9M
 
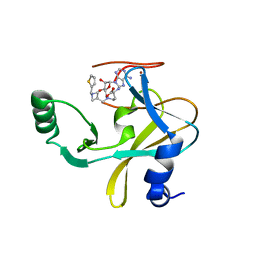 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
9C4M
 
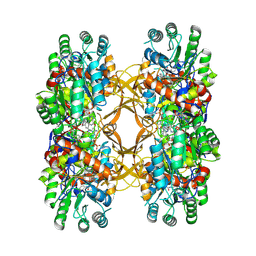 | | Crystal Structure of A. baumannii GuaB dCBS with inhibitor G6 (3826) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(morpholin-4-yl)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Differential effects of inosine monophosphate dehydrogenase (IMPDH/GuaB) inhibition in Acinetobacter baumannii and Escherichia coli.
J.Bacteriol., 2024
|
|
6UCF
 
 | |
7WVH
 
 | |
8BNS
 
 | |
7X31
 
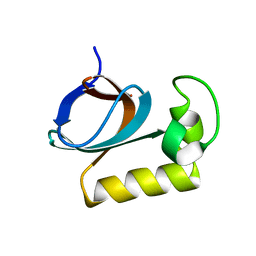 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
6B41
 
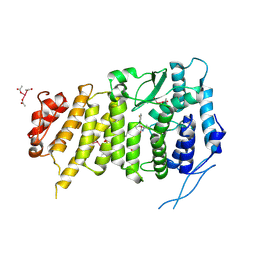 | | Menin bound to M-525 | | Descriptor: | Menin, methyl {(1S,2R)-2-[(S)-cyano[1-({1-[4-({1-[4-(dimethylamino)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl](3-fluorophenyl)methyl]cyclopentyl}carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8K60
 
 | |
6MPH
 
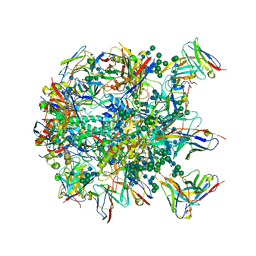 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
