7DK9
 
 | |
7CVH
 
 | | Human Fructose-1,6-bisphosphatase 1 in complex with geranylgeranyl diphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structural basis for GGPP activation on the enzymatic activities FBP1
To Be Published
|
|
7WVB
 
 | | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state | | Descriptor: | Fructose-1,6-bisphosphatase 1 | | Authors: | Chen, Y, Zhang, J, Li, C, Cao, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Human Fructose-1,6-bisphosphatase 1 mutant R50A in APO R-state
To Be Published
|
|
7X7H
 
 | | Crystal structure of Fructose regulator/Histidine phosphocarrier protein complex from Vibrio cholerae | | Descriptor: | CALCIUM ION, Catabolite repressor/activator, HPr family phosphocarrier protein | | Authors: | Kim, M.-K, Zhang, J, Yoon, C.-K, Seok, Y.-J. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HPr prevents FruR-mediated facilitation of RNA polymerase binding to the fru promoter in Vibrio cholerae.
Nucleic Acids Res., 51, 2023
|
|
7XIO
 
 | | Crystal structure of TYR from Ralstonia | | Descriptor: | PHOSPHATE ION, Polyphenol oxidase | | Authors: | Sun, D.Y, Cui, P.P, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Zhang, J, Li, X, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of TYR from Ralstonia
To Be Published
|
|
7YGF
 
 | | Crystal structure of YggS from Fusobacterium nucleatum | | Descriptor: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | He, S.R, Chan, Y.Y, Wang, L.L, Bai, X, Bu, T.T, Zhang, J, Xu, Y.B. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum.
Molecules, 27, 2022
|
|
7AMN
 
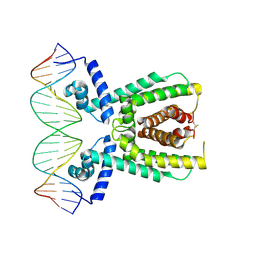 | | Structure of LuxR with DNA (repression) | | Descriptor: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
7AMT
 
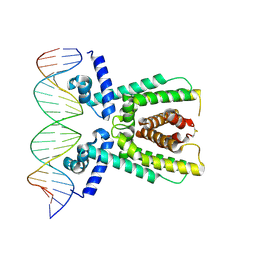 | | Structure of LuxR with DNA (activation) | | Descriptor: | DNA (5'-D(P*AP*TP*AP*AP*TP*GP*AP*CP*AP*TP*TP*AP*CP*TP*GP*TP*AP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*AP*GP*TP*AP*AP*TP*GP*TP*CP*AP*TP*TP*AP*T)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
2XVO
 
 | | SSO1725, a protein involved in the CRISPR/Cas pathway | | Descriptor: | BETA-MERCAPTOETHANOL, SSO1725, SULFATE ION | | Authors: | Reeks, J, Liu, H, Naismith, J, White, M, McMahon, S. | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-29 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mechanism of the Cmr Complex for Crispr-Mediated Antiviral Immunity.
Mol.Cell, 45, 2012
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
3DJ3
 
 | |
3DIW
 
 | | c-terminal beta-catenin bound TIP-1 structure | | Descriptor: | Tax1-binding protein 3, decameric peptide form Catenin beta-1 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
3DJ1
 
 | | crystal structure of TIP-1 wild type | | Descriptor: | SULFATE ION, Tax1-binding protein 3 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1.
J.Mol.Biol., 384, 2008
|
|
5DIJ
 
 | | The crystal structure of CT | | Descriptor: | CHLORIDE ION, GLYCEROL, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
5DLK
 
 | | The crystal structure of CT mutant | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, TqaA | | Authors: | Zhang, J.R, Tang, Y, Zhou, J.H. | | Deposit date: | 2015-09-06 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of nonribosomal peptide macrocyclization in fungi
Nat.Chem.Biol., 12, 2016
|
|
6RX8
 
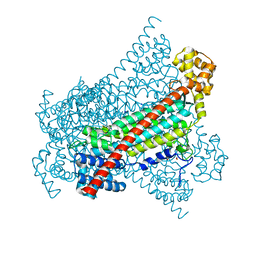 | |
6RXA
 
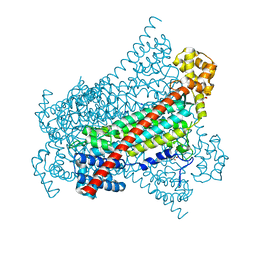 | | EDDS lyase variant D290M/Y320M with bound formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, GLYCEROL, ... | | Authors: | Grandi, E, Poelarends, G.J, Thunnissen, A.M.W.H. | | Deposit date: | 2019-06-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Engineered C-N Lyase: Enantioselective Synthesis of Chiral Synthons for Artificial Dipeptide Sweeteners.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8AG1
 
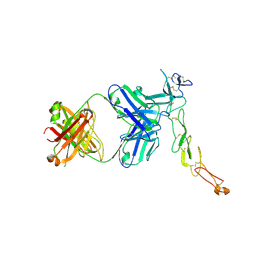 | | Crystal structure of a novel OX40 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Novel OX40 antibody heavy chain, Novel OX40 antibody light chain, ... | | Authors: | Gao, H, Zhou, A. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structural Basis of a Novel Agonistic Anti-OX40 Antibody.
Biomolecules, 12, 2022
|
|
8HIP
 
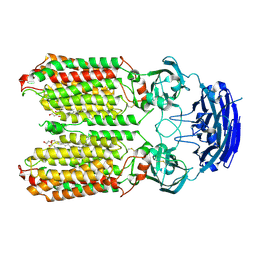 | | dsRNA transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Systemic RNA interference defective protein 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HKE
 
 | | dsRNA transporter | | Descriptor: | RNA (37-MER), Systemic RNA interference defective protein 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4NSO
 
 | | Crystal structure of the effector-immunity protein complex | | Descriptor: | Effector protein, Immunity protein | | Authors: | Dong, C. | | Deposit date: | 2013-11-28 | | Release date: | 2014-04-16 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recognition of the type VI spike protein VgrG3 by a cognate immunity protein.
Febs Lett., 588, 2014
|
|
4NSR
 
 | |
8HKK
 
 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
