6VH6
 
 | |
9DI9
 
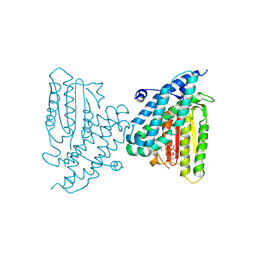 | | Rat branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (3M)-6-fluoro-3-(2,4,5-trifluoro-3-methoxyphenyl)-1-benzothiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, Branched-chain alpha-ketoacid dehydrogenase kinase, ... | | Authors: | Liu, S. | | Deposit date: | 2024-09-05 | | Release date: | 2024-11-27 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Discovery of First Branched-Chain Ketoacid Dehydrogenase Kinase (BDK) Inhibitor Clinical Candidate PF-07328948.
J.Med.Chem., 68, 2025
|
|
8QBN
 
 | |
8QE8
 
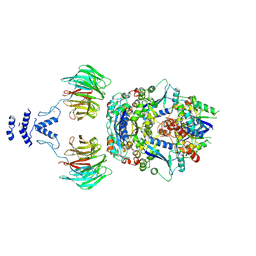 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
7JOU
 
 | |
6TZP
 
 | | W96F Oxalate Decarboxylase (B. subtilis) | | Descriptor: | MANGANESE (II) ION, Oxalate decarboxylase | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-08-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
7JOV
 
 | |
7NB4
 
 | | Structure of Mcl-1 complex with compound 1 | | Descriptor: | (2~{R})-2-[[5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
7NB7
 
 | | Structure of Mcl-1 complex with compound 6b | | Descriptor: | (2~{R})-2-[[7-but-2-ynyl-5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-pyrrolo[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
8F5J
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | 3-chloro-5-fluorothieno[3,2-b]thiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
8F5F
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2P)-2-[(4P)-4-{6-[(1-ethylcyclopropyl)methoxy]pyridin-3-yl}-1,3-thiazol-2-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
8F5S
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2M)-2-[2-(4-methylphenyl)-1,3-thiazol-4-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
2H2Q
 
 | | Crystal structure of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Senkovich, O, Schormann, N, Chattopadhyay, D. | | Deposit date: | 2006-05-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
8P2M
 
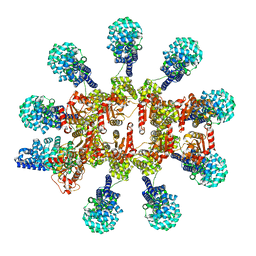 | | C. elegans TIR-1 protein. | | Descriptor: | NAD(+) hydrolase tir-1 | | Authors: | Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans.
Cell Rep, 42, 2023
|
|
8S41
 
 | | The structure of the copia retrotransposon icosahedral capsid (T=9) | | Descriptor: | Copia VLP protein | | Authors: | Klumpe, S, Beck, F, Briggs, J.A.G, Beck, M, Plitzko, J.M. | | Deposit date: | 2024-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | In-cell structure and snapshots of copia retrotransposons in intact tissue by cryo-ET.
Cell, 188, 2025
|
|
9FH0
 
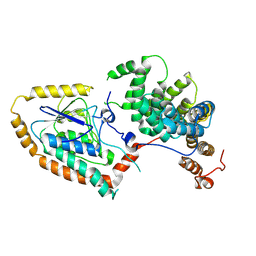 | | Pex5-Eci1 complex - Pex5 local refinement | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, Peroxisomal targeting signal receptor | | Authors: | Elad, N, Dym, O. | | Deposit date: | 2024-05-26 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An unconventional interaction interface between the peroxisomal targeting factor Pex5 and Eci1 enables PTS1 independent import
To Be Published
|
|
9FGZ
 
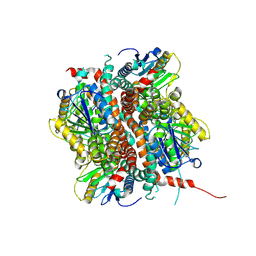 | | Pex5-Eci1 complex - Eci1 reconstruction | | Descriptor: | 3,2-trans-enoyl-CoA isomerase | | Authors: | Elad, N, Dym, O. | | Deposit date: | 2024-05-26 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An unconventional interaction interface between the peroxisomal targeting factor Pex5 and Eci1 enables PTS1 independent import
To Be Published
|
|
8P2L
 
 | |
2GIW
 
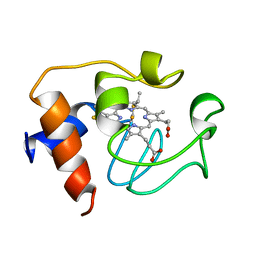 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, 40 STRUCTURES | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-25 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
1KBG
 
 | | MHC Class I H-2KB Presented Glycopeptide RGY8-6H-GAL2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-2-MICROGLOBULIN), ... | | Authors: | Speir, J.A, Abdel-Motal, U.M, Jondal, M, Wilson, I.A. | | Deposit date: | 1998-08-28 | | Release date: | 1999-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an MHC class I presented glycopeptide that generates carbohydrate-specific CTL.
Immunity, 10, 1999
|
|
6UL7
 
 | | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Gardner, S, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-10-07 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole
To be Published
|
|
9BPX
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (1'-(4-((1-butylpyrrolidin-3-yl)methoxy)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, HISTIDINE, [(1'R)-1'-(4-{[(3R)-1-(3-fluoropropyl)pyrrolidin-3-yl]methoxy}phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone, ... | | Authors: | Young, K.Y, Fanning, S.W. | | Deposit date: | 2024-05-08 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting unique ligand binding domain structural features downregulates DKK1 in Y537S ESR1 mutant breast cancer cells.
Breast Cancer Res, 27, 2025
|
|
1Q1N
 
 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-07-22 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
1PIW
 
 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-05-30 | | Release date: | 2004-08-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
1PS0
 
 | | Crystal Structure of the NADP(H)-Dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-06-20 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
