6YGN
 
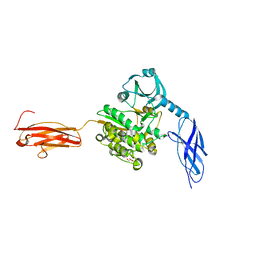 | | Titin kinase and its flanking domains | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Titin | | Authors: | Fleming, J.R, Franke, B, Bogomolovas, J, Mayans, O. | | Deposit date: | 2020-03-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Titin kinase ubiquitination aligns autophagy receptors with mechanical signals in the sarcomere.
Embo Rep., 22, 2021
|
|
6Z4K
 
 | |
6Z4J
 
 | |
6Z4M
 
 | |
6Z4G
 
 | |
6Z3V
 
 | |
7P6X
 
 | | Cryo-Em structure of the hexameric RUVBL1-RUVBL2 in complex with ZNHIT2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Llorca, O, Serna, M, Fernandez-Leiro, R. | | Deposit date: | 2021-07-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of RUVBL1-RUVBL2-ZNHIT2, a complex that interacts with pre-mRNA-processing-splicing factor 8.
Nucleic Acids Res., 50, 2022
|
|
6Z4I
 
 | |
6Z4O
 
 | |
8F5V
 
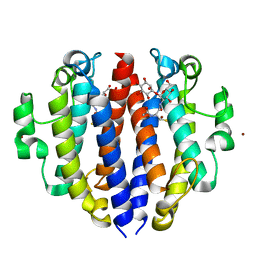 | |
6Z4H
 
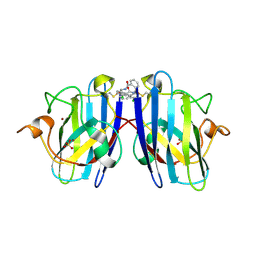 | |
6Z4L
 
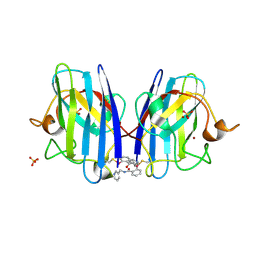 | |
4LVC
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenosine | | Descriptor: | ACETATE ION, ADENOSINE, AMMONIUM ION, ... | | Authors: | Manszewski, T, Singh, K, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An enzyme captured in two conformational states: crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6FE6
 
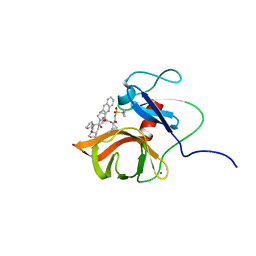 | | Solution structure of a last generation P2-P4 macrocyclic inhibitor | | Descriptor: | (3aR,7S,10S,12R,24aR)-7-cyclopentyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-5,8-dioxo-1,2,3,3a,5,6,7,8,11,12,20,21,22,23,24,24a-hexadecahydro-10H-9,12-methanocyclopenta[18,19][1,10,3,6]dioxadiazacyclononadecino[12,11-b]quinoline-10-carboxamide, Non-structural 3 protease, ZINC ION | | Authors: | Gallo, M, Eliseo, T, Cicero, D.O. | | Deposit date: | 2017-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a last generation macrocyclic inhibitor. Hepatitis C virus NS3 protease complex: when S prime region occupancy is not enough to stabilize the protein conformation in the absence of NS4A.
To Be Published
|
|
3FZD
 
 | | Mutation of Asn28 disrupts the enzymatic activity and dimerization of SARS 3CLpro | | Descriptor: | 3C-like proteinase | | Authors: | Barrila, J, Gabelli, S, Bacha, U, Amzel, L.M, Freire, E. | | Deposit date: | 2009-01-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CL(pro) .
Biochemistry, 49, 2010
|
|
3PI5
 
 | | Crystal Structure of Human Beta Secretase in Complex with BFG356 | | Descriptor: | (3S,4S,5R)-3-(3-bromo-4-hydroxybenzyl)-5-[(3-cyclopropylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-23 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6HPV
 
 | | Crystal structure of mouse fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Fetuin-B | | Authors: | Fahrenkamp, D, Dietzel, E, de Sanctis, D, Jovine, L. | | Deposit date: | 2018-09-22 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
1G96
 
 | | HUMAN CYSTATIN C; DIMERIC FORM WITH 3D DOMAIN SWAPPING | | Descriptor: | CHLORIDE ION, CYSTATIN C, GLYCEROL | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Grubb, A, Abrahamson, M, Jaskolski, M. | | Deposit date: | 2000-11-22 | | Release date: | 2001-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping.
Nat.Struct.Biol., 8, 2001
|
|
2RSC
 
 | |
5W4G
 
 | |
2YTX
 
 | | Solution structure of the second cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | Cold shock domain-containing protein E1 | | Authors: | Goroncy, A.K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2YUX
 
 | | Solution Structure of 3rd Fibronectin type three Domain of slow type Myosin-Binding Protein C | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Niraula, T.N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 3rd Fibronectin type three Domain of slow type Myosin-Binding Protein C
To be Published
|
|
5G0T
 
 | | InhA in complex with a DNA encoded library hit | | Descriptor: | 1-benzyl-N-[cis-4-(2-{[(4-fluorophenyl)methyl][2-(methylamino)-2-oxoethyl]amino}-2-oxoethyl)cyclohexyl]-5-methyl-1H-1,2,3-triazole-4-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of Cofactor-Specific, Bactericidal Mycobacterium Tuberculosis Inha Inhibitors Using DNA-Encoded Library Technology
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G0U
 
 | | InhA in complex with a DNA encoded library hit | | Descriptor: | 5-[(4-fluoranyl-3-phenoxy-phenyl)methylamino]-~{N}-methyl-6-[(1-pyridin-2-ylpiperidin-4-yl)amino]pyridine-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Cofactor-Specific, Bactericidal Mycobacterium Tuberculosis Inha Inhibitors Using DNA-Encoded Library Technology
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3FKR
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase complex with pyruvate | | Descriptor: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION, SODIUM ION | | Authors: | Shimada, N, Mikami, B. | | Deposit date: | 2008-12-17 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
