6UK6
 
 | |
6UKB
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-[5-(4-{[5-(acetylamino)-1,3,4-thiadiazol-2-yl]oxy}piperidin-1-yl)-1,3,4-thiadiazol-2-yl]acetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00020
To Be Published
|
|
6ULA
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-cyclopropyl-N-{5-[4-({5-[(cyclopropylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00030
To Be Published
|
|
6UJM
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00013 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-{5-[(3R)-3-{[5-(acetylamino)-1,3,4-thiadiazol-2-yl]amino}pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00013
To Be Published
|
|
6UL9
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00023 | | Descriptor: | 2-phenyl-N-{5-[(1-{5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}azetidin-3-yl)oxy]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q.Q, Cerione, R.A. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
8XD7
 
 | |
8XDB
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | (5E)-2-azanylidene-5-[(2,3-dimethoxyphenyl)methylidene]-1,3-thiazolidin-4-one, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDC
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | N-[3-[1,1-bis(oxidanylidene)-1,2-thiazolidin-2-yl]-4-chloranyl-phenyl]-2-methoxy-5-methyl-benzenesulfonamide, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDI
 
 | | Cryo-EM structure of zebrafish urea transporter. | | Descriptor: | N-(4-acetamidophenyl)-5-ethanoyl-furan-2-carboxamide, Urea transporter | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
5ZTC
 
 | |
6XEO
 
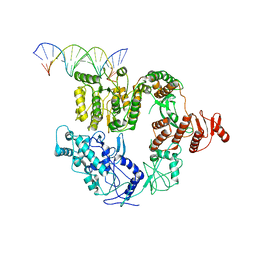 | | Structure of Mfd bound to dsDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | Authors: | Brugger, C, Deaconescu, A. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
8T8M
 
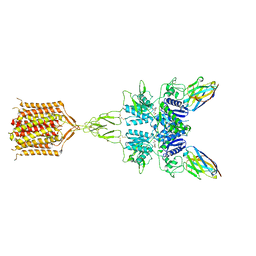 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nb43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-22 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T6J
 
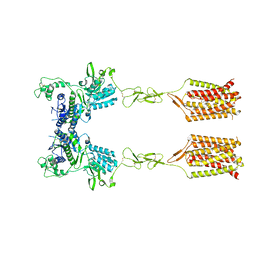 | | CDPPB-bound inactive mGlu5 | | Descriptor: | 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8TAO
 
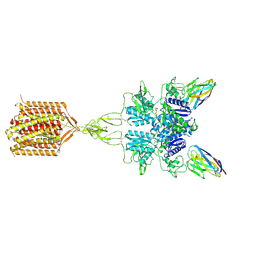 | | Quis and CDPPB bound active mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5, ... | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T7H
 
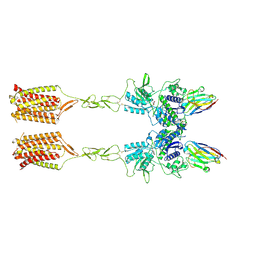 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-20 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8HGI
 
 | | Crystal structure of VNAR aGFP14 in complex with GFP | | Descriptor: | GFP, VNAR aGFP14 | | Authors: | Zheng, P, Zhu, C, Jin, T. | | Deposit date: | 2022-11-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selection, identification and crystal structure of shark-derived single-domain antibodies against a green fluorescent protein.
Int.J.Biol.Macromol., 247, 2023
|
|
7U1Z
 
 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
8I88
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | Descriptor: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8I87
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Zhang, H, Deng, Z.Q, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
7YSX
 
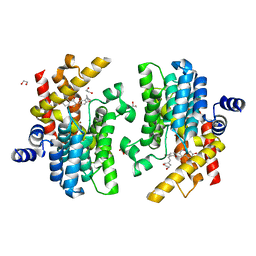 | | Crystal structure of PDE4D complexed with licoisoflavone A | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YQF
 
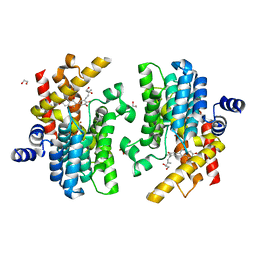 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GDB
 
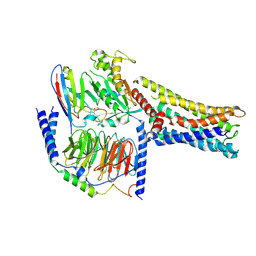 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jianqiang, M, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GDC
 
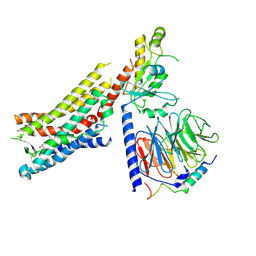 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 3 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Yang, D, Shiyi, G, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8W56
 
 | | Cryo-EM structure of DSR2-DSAD1 state 1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-08-25 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
5WWO
 
 | | Crystal structure of Enp1 | | Descriptor: | Essential nuclear protein 1, Protein LTV1 | | Authors: | Ye, K, Zhang, W. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
