4MKC
 
 | | Crystal Structure of Anaplastic Lymphoma Kinase Complexed with LDK378 | | Descriptor: | 5-chloro-N~2~-[5-methyl-4-(piperidin-4-yl)-2-(propan-2-yloxy)phenyl]-N~4~-[2-(propan-2-ylsulfonyl)phenyl]pyrimidine-2,4-diamine, ALK tyrosine kinase receptor, GLYCEROL | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2013-09-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The ALK Inhibitor Ceritinib Overcomes Crizotinib Resistance in Non-Small Cell Lung Cancer.
Cancer Discov, 4, 2014
|
|
4JML
 
 | |
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
7JLT
 
 | | Crystal Structure of SARS-CoV-2 NSP7-NSP8 complex. | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Biswal, M, Hai, R, Song, J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP.
Nucleic Acids Res., 49, 2021
|
|
4Q44
 
 | | Polymerase-Damaged DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*TP*CP*TP*AP*(RDG)P*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA polymerase IV, ... | | Authors: | Nair, D.T, Kottur, J, Sharma, A. | | Deposit date: | 2014-04-13 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
1TPB
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1TPC
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
7YL8
 
 | |
6HU9
 
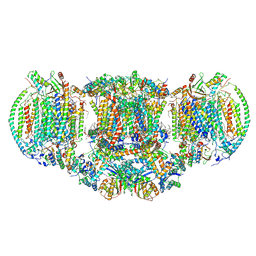 | | III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CALCIUM ION, ... | | Authors: | Hartley, A.M, Pinotsis, N, Marechal, A. | | Deposit date: | 2018-10-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5UG6
 
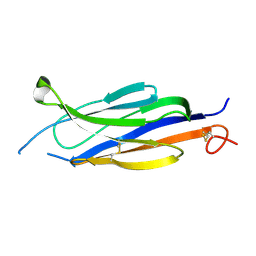 | | Perforin C2 Domain - T431D | | Descriptor: | IODIDE ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
3NSJ
 
 | | The X-ray crystal structure of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Law, R.H, Whisstock, J.C, Caradoc-Davies, T.T. | | Deposit date: | 2010-07-01 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structural basis for membrane binding and pore formation by lymphocyte perforin.
Nature, 468, 2010
|
|
1REK
 
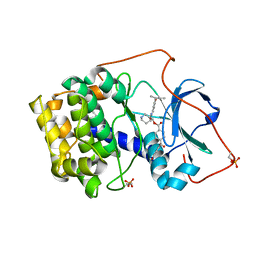 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 8 | | Descriptor: | 3-[(3-SEC-BUTYL-4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXY-5-METHOXYBENZOYL)BENZOATE, PENTANAL, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1RE8
 
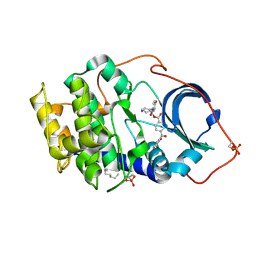 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 2 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXYBENZOYL)BENZOATE, N-OCTANE, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
4IR9
 
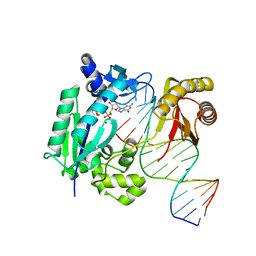 | | Polymerase-DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(P*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Sharma, A, Nair, D.T. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IRD
 
 | | Structure of Polymerase-DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IR1
 
 | | Polymerase-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Sharma, A, Nair, D.T. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
1REJ
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-HYDROXYBENZOATE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
3GZQ
 
 | | HUMAN SOD1 A4V Metal-free Variant | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Galaleldeen, A, Taylor, A.B, Whitson, L.J, Hart, P.J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A.
Arch.Biochem.Biophys., 492, 2009
|
|
4IRC
 
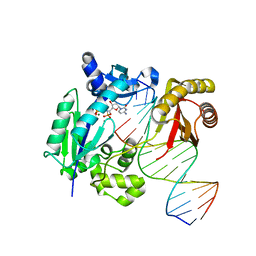 | | Polymerase-DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IRK
 
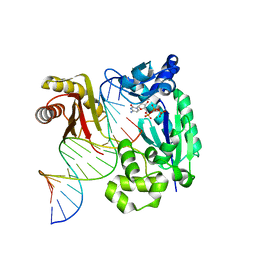 | | structure of Polymerase-DNA complex, dna | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*A*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3'), DNA (5'-D(*TP*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
3GZP
 
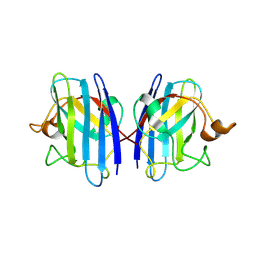 | |
7EIL
 
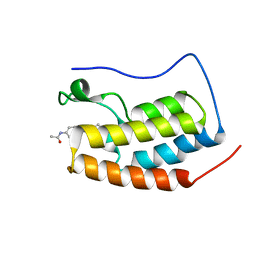 | | BRD4-BD1 in complex with LT-909-110 | | Descriptor: | Bromodomain-containing protein 4, N-[4-[4-ethanoyl-5-methyl-2-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-pyrrol-3-yl]phenyl]ethanamide | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 1-(5-(1H-benzo[d]imidazole-2-yl)-2,4-dimethyl-1H-pyrrol-3-yl)ethan-1-one derivatives as novel and potent bromodomain and extra-terminal (BET) inhibitors with anticancer efficacy
Eur.J.Med.Chem., 227, 2022
|
|
8GVM
 
 | | The structure of azide-bound cytochrome C oxidase determined using the crystals exposed to 20 mm azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2022-09-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity
J. Biol. Chem., 293, 2018
|
|
3W56
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
3W57
 
 | | Structure of a C2 domain | | Descriptor: | C2 domain protein, CALCIUM ION | | Authors: | Traore, D.A.K, Whisstock, J.C. | | Deposit date: | 2013-01-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Defining the interaction of perforin with calcium and the phospholipid membrane.
Biochem.J., 456, 2013
|
|
