4HGL
 
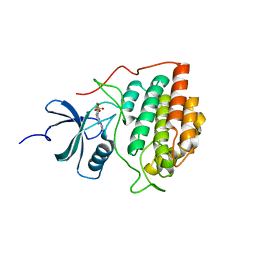 | | Crystal structure of ck1g3 with compound 1 | | Descriptor: | 2-[5-methoxy-2-(quinolin-3-yl)pyrimidin-4-yl]-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform gamma-3, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Potent and Selective CK1 gamma Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
6YQG
 
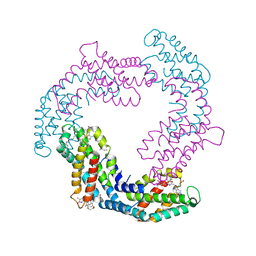 | | Crystal structure of native Phycocyanin in spacegroup P63 at 1.45 Angstroms. | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-17 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6Y6O
 
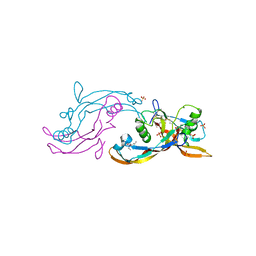 | | Structure of mature activin A with small molecule 42 | | Descriptor: | (3~{R})-4-ethyl-3-methyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
5J7A
 
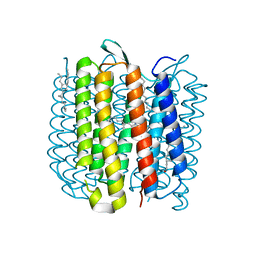 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
6Y6N
 
 | | Structure of mature activin A with small molecule 2 | | Descriptor: | (3~{R})-3,4-dimethyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6YPQ
 
 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup R32 at 1.29 Angstroms | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, GLYCINE, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6YQ8
 
 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P63 at 1.8 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6YYJ
 
 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P21212 at 2.1 Angstroms | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, C-phycocyanin alpha chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5XFD
 
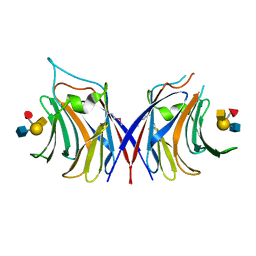 | |
6YN0
 
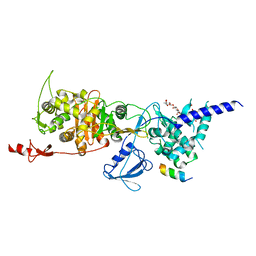 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | Descriptor: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
5AH4
 
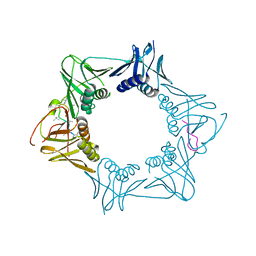 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | Descriptor: | CYCLOHEXYL GRISELIMYCIN, DNA POLYMERASE III SUBUNIT BETA, SODIUM ION | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
5X7E
 
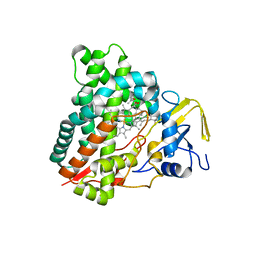 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D2 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(E,2R,5S)-5,6-dimethyl-6-oxidanyl-hept-3-en-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydr o-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D3 dihydroxylase | | Authors: | Hayashi, K, Yasuda, K, Shiro, Y, Sugimoto, H, Sakaki, T. | | Deposit date: | 2017-02-25 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Production of an active form of vitamin D2 by genetically engineered CYP105A1
Biochem. Biophys. Res. Commun., 486, 2017
|
|
4OD2
 
 | |
4WYV
 
 | |
5AGU
 
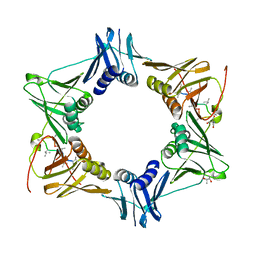 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA POLYMERASE III SUBUNIT BETA, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
5AH2
 
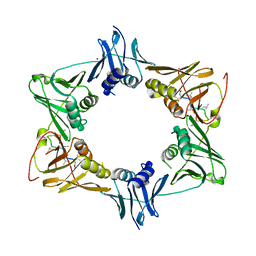 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | Descriptor: | DNA POLYMERASE III SUBUNIT BETA, GRISELIMYCIN, SODIUM ION | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
4WNX
 
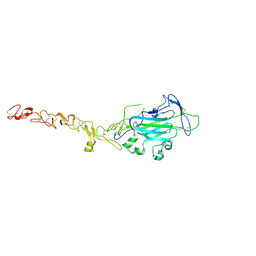 | | Netrin 4 lacking the C-terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McDougall, M, Patel, T, Reuten, R, Meier, M, Koch, M, Stetefeld, J. | | Deposit date: | 2014-10-14 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes.
Nat Commun, 7, 2016
|
|
4N4V
 
 | |
4N3R
 
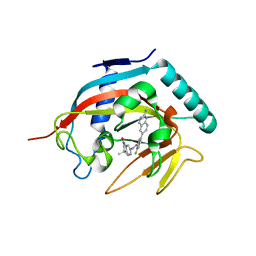 | | Co-crystal structure of tankyrase 1 with compound 2 (5-(2-aminoquinazolin-6-yl)-N-(4,4-dimethyl-2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)-2-fluorobenzamide) | | Descriptor: | 5-(2-aminoquinazolin-6-yl)-N-(4,4-dimethyl-2-oxo-1,2,3,4-tetrahydroquinolin-7-yl)-2-fluorobenzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2013-10-07 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of 2-aminopyridine oxazolidinones as potent and selective tankyrase inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
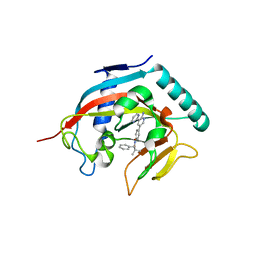
4N4T
 
 | |
1WD5
 
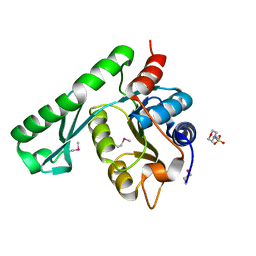 | | Crystal structure of TT1426 from Thermus thermophilus HB8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, hypothetical protein TT1426 | | Authors: | Shibata, R, Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-11 | | Release date: | 2004-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution
Protein Sci., 14, 2005
|
|
3HJR
 
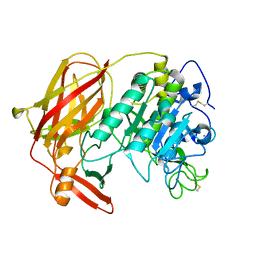 | | Crystal structure of serine protease of Aeromonas sobria | | Descriptor: | CALCIUM ION, Extracellular serine protease | | Authors: | Utsunomiya, H, Tsuge, H, Kobayashi, H, Okamoto, K. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the kexin-like serine protease from Aeromonas sobria as a sepsis-causing factor
J.Biol.Chem., 284, 2009
|
|
1DMK
 
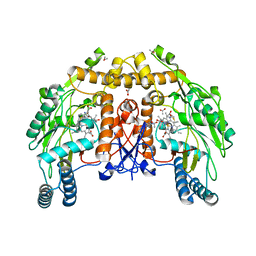 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
1YJI
 
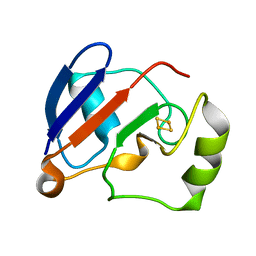 | | RDC-refined Solution NMR structure of reduced putidaredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Jain, N.U, Tjioe, E, Savidor, A, Boulie, J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent structural differences in putidaredoxin derived from homologous structure refinement via residual dipolar couplings.
Biochemistry, 44, 2005
|
|
