8P9C
 
 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | 1,2-ETHANEDIOL, Darpin 1810 F11, Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
8P9D
 
 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 A2 | | Descriptor: | Darpin 1810 A2, Tumor protein 63, Tumor protein p73 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
7ZEJ
 
 | | Crystal structure of the human MGC45594 gene product in complex with celecoxib. | | Descriptor: | 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 3 | | Authors: | Shafqat, N, Yue, W.W, Koekemoer, L, Niesen, F, Ugochukwu, E, Vollmar, M, Weigelt, J, Krojer, T, Pike, A, Chaikaud, A, Von Delft, F, Arrowsmith, C, Bountra, C, Edwards, A, Opperman, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Human prostaglandin/alkenal reductases: substrate specificities, inhibitor profiles, structural insights and subcellular localization suggest protective roles in inflammatory and oxidative stress conditions.
To Be Published
|
|
7Q8V
 
 | | Crystal structure of TTBK1 in complex with VNG2.73 (compound 42) | | Descriptor: | PHOSPHATE ION, Tau-tubulin kinase 1, ~{N}-[4-(2-chloranylphenoxy)phenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-amine | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7Q8W
 
 | | Crystal structure of TTBK1 in complex with VNG1.35 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Tau-tubulin kinase 1, ... | | Authors: | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
6OAW
 
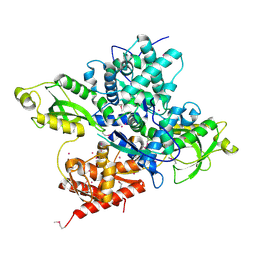 | | Crystal structure of a CRISPR Cas-related protein | | Descriptor: | UNKNOWN ATOM OR ION, WYL1 | | Authors: | Zhang, H, Dong, C, Li, L, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the modulatory role of the accessory protein WYL1 in the Type VI-D CRISPR-Cas system.
Nucleic Acids Res., 47, 2019
|
|
6OGK
 
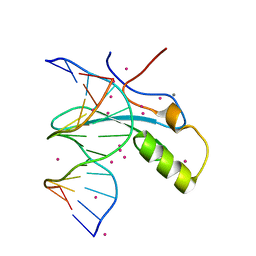 | | MeCP2 MBD in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
7UFV
 
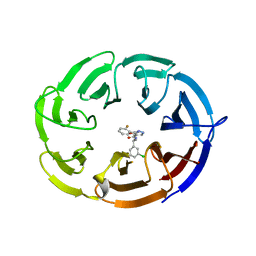 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
4DO0
 
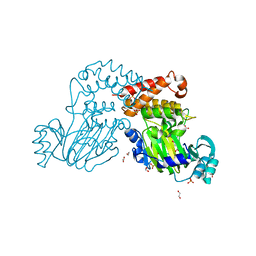 | | Crystal Structure of human PHF8 in complex with Daminozide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DAMINOZIDE, ... | | Authors: | Krojer, T, Daniel, M, Ng, S.S, Walport, L.J, Chowdhury, R, Arrowsmith, C.H, Edwards, A, Bountra, C, Kawamura, A, Muller-Knapp, S, McDonough, M.A, von Delft, F, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: |
|
|
6ICV
 
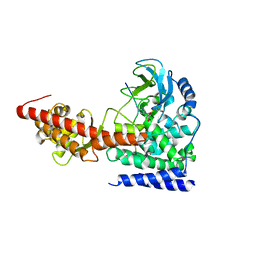 | | Structure of SETD3 bound to SAH and unmodified actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
3BKB
 
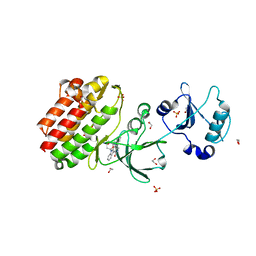 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES) | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, ... | | Authors: | Filippakopoulos, P, Salah, E, Fedorov, O, Cooper, C, Ugochukwu, E, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
6FCX
 
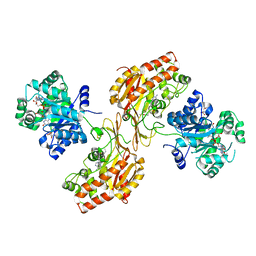 | | Structure of human 5,10-methylenetetrahydrofolate reductase (MTHFR) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase, ... | | Authors: | Kopec, J, Bezerra, G.A, Oberholzer, A.E, Rembeza, E, Sorrell, F.J, Chalk, R, Borkowska, O, Ellis, K, Kupinska, K, Krojer, T, Burgess-Brown, N, Von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Froese, D.S, Baumgartner, M, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-21 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition.
Nat Commun, 9, 2018
|
|
6V3N
 
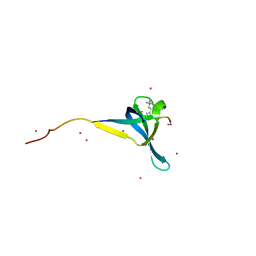 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
8BI5
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0737 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-~{N}-methyl-~{N}-(1-propan-2-ylpiperidin-4-yl)-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0737
To Be Published
|
|
8BI6
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0638 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0638
To Be Published
|
|
7MWI
 
 | | Crystal structure of human BAZ2A | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
6V8W
 
 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6QAT
 
 | | Crystal structure of ULK2 in complexed with hesperadin | | Descriptor: | N-{(3Z)-2-oxo-3-[phenyl({4-[(piperidin-1-yl)methyl]phenyl}amino)methylidene]-2,3-dihydro-1H-indol-5-yl}ethanesulfonamide, Serine/threonine-protein kinase ULK2 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6ICT
 
 | | Structure of SETD3 bound to SAH and methylated actin | | Descriptor: | Actin, cytoplasmic 1, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Guo, Q, Liao, S, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-07 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural insights into SETD3-mediated histidine methylation on beta-actin.
Elife, 8, 2019
|
|
6QAS
 
 | | Crystal structure of ULK1 in complexed with PF-03814735 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
6QAU
 
 | | Crystal structure of ULK2 in complexed with MRT67307 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
8PP0
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with JP147 | | Descriptor: | 3-[4-[2,3-dihydro-1H-inden-4-yl(methyl)amino]-6-(trifluoromethyl)pyrimidin-2-yl]oxypropanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of a Highly Potent Partial RXR Agonist with Superior Physicochemical Properties.
J.Med.Chem., 67, 2024
|
|
8QZ2
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an inhibitory nanobody (Nb61) | | Descriptor: | Nanobody 61, POTASSIUM ION, Potassium channel subfamily K member 10 | | Authors: | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
6QAV
 
 | | Crystal structure of ULK2 in complexed with MRT68921 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SODIUM ION, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Biochem.J., 476, 2019
|
|
8QZ1
 
 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with a nanobody (Nb58) | | Descriptor: | Isoform B of Potassium channel subfamily K member 10, Nanobody 58, POTASSIUM ION | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Baronina, A, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.588 Å) | | Cite: | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
