8OW3
 
 | | Crystal structure of wild-type c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OV7
 
 | | Crystal structure of D1228V c-MET bound by compound 10 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-[3-(1H-imidazol-5-yl)phenyl]ethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8R3D
 
 | | Crystal structure of Pent only at pH 8.8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta propeller | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-11-08 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Multivalent Calixarene Complexation of a Designed Pentameric Lectin.
Biomacromolecules, 25, 2024
|
|
7QQB
 
 | | Crystal structure of the envelope glycoprotein complex of Puumala virus in complex with the scFv fragment of the broadly neutralizing human antibody ADI-42898 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, GLYCEROL, ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2022-01-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic basis of neutralization by a pan-hantavirus protective antibody.
Sci Transl Med, 15, 2023
|
|
8RIH
 
 | | Crystal structure of the Saccharomyces cerevisiae URH1p riboside hydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Uridine ribohydrolase | | Authors: | Degano, M, Carriles, A.A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure-Function Insights into the Dual Role in Nucleobase and Nicotinamide Metabolism and a Possible Use in Cancer Gene Therapy of the URH1p Riboside Hydrolase.
Int J Mol Sci, 25, 2024
|
|
8R3C
 
 | |
8VWW
 
 | | CCHFV GP38 bound to ADI-46152 and ADI-58048 Fabs | | Descriptor: | ADI-46152 Fab Heavy Chain, ADI-46152 Fab Light Chain, ADI-58048 Fab Heavy Chain, ... | | Authors: | Hjorth, C.K, McLellan, J.S. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Cell Rep, 43, 2024
|
|
8VVK
 
 | | CCHFV GP38 bound to ADI-46143 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-46143 Fab Heavy Chain, ADI-46143 Fab Light Chain, ... | | Authors: | Hjorth, C.K, McLellan, J.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Cell Rep, 43, 2024
|
|
8VVL
 
 | | CCHFV GP38 bound to c13G8 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hjorth, C.K, Mishra, A.K, McLellan, J.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Cell Rep, 43, 2024
|
|
3PGA
 
 | | STRUCTURAL CHARACTERIZATION OF PSEUDOMONAS 7A GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Ammon, H.L, Copeland, T.D, Swain, A.L. | | Deposit date: | 1994-07-19 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Pseudomonas 7A glutaminase-asparaginase.
Biochemistry, 33, 1994
|
|
6HQU
 
 | | Humanised RadA mutant HumRadA22 in complex with a recombined BRC repeat 8-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Breast cancer type 2 susceptibility, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Lindenburg, L, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Improved RAD51 binders through motif shuffling based on the modularity of BRC repeats.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EXB
 
 | |
5EXC
 
 | |
3OKA
 
 | | Crystal structure of Corynebacterium glutamicum PimB' in complex with GDP-Man (triclinic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
3OKC
 
 | | Crystal structure of Corynebacterium glutamicum PimB' bound to GDP (orthorhombic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
5T1T
 
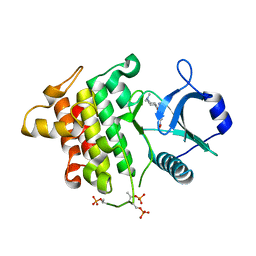 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(6-nitroquinazolin-4-yl)oxy-cyclohexan-1-amine | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5T1S
 
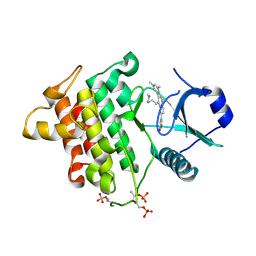 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | 5-[3-(3,5-dimethylphenyl)-4-[4-(methylamino)butyl]quinolin-6-yl]pyridin-3-ol, Interleukin-1 receptor-associated kinase 4 | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3OKP
 
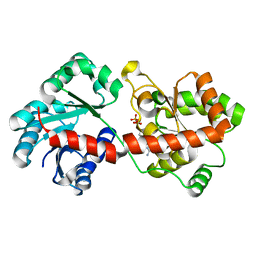 | | Crystal structure of Corynebacterium glutamicum PimB' bound to GDP-Man (orthorhombic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
8AQF
 
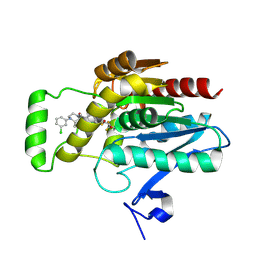 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND LEI-515 | | Descriptor: | 1-[(~{R})-[2-chloranyl-4-[(2~{S},3~{S})-4-(3-chlorophenyl)-2,3-dimethyl-piperazin-1-yl]carbonyl-phenyl]sulfinyl]-3,3-bis(fluoranyl)pentan-2-one, Monoglyceride lipase | | Authors: | Jiang, M, Huizenga, M, Wirt, J, Paloczi, J, Amedi, A, van der Berg, R, Benz, J, Collin, L, Deng, H, Driever, W, Florea, B, Grether, U, Janssen, A, Heitman, L, Lam, T.W, Mohr, F, Pavlovic, A, Ruf, I, Rutjes, H, Stevens, F, van der Vliet, D, van der Wel, T, Wittwer, M, Boeckel, C, Pacher, P, Hohmann, A, van der Stelt, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a peripheral restricted, reversible monoacylglycerol lipase inhibitor that reduces liver injury and chemotherapy-induced neuropathy
To Be Published
|
|
2PSZ
 
 | |
2PT0
 
 | |
4NZ0
 
 | |
4NYZ
 
 | | The EMCV 3Dpol structure with altered motif A conformation at 2.15A resolution | | Descriptor: | GLUTAMINE, GLYCEROL, Genome polyprotein, ... | | Authors: | Vives-adrian, L, Lujan, C, Ferrer-orta, C, Verdaguer, N. | | Deposit date: | 2013-12-11 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site
J.Virol., 88, 2014
|
|
1TXI
 
 | | Crystal structure of the vdr ligand binding domain complexed to TX522 | | Descriptor: | (1R,3R)-5-((Z)-2-((1R,7AS)-HEXAHYDRO-1-((S)-6-HYDROXY-6-METHYLHEPT-4-YN-2-YL)-7A-METHYL-1H-INDEN-4(7AH)-YLIDENE)ETHYLIDENE)CYCLOHEXANE-1,3-DIOL, Vitamin D receptor | | Authors: | Moras, D, Rochel, N. | | Deposit date: | 2004-07-05 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Superagonistic Action of 14-epi-Analogs of 1,25-Dihydroxyvitamin D Explained by Vitamin D Receptor-Coactivator Interaction
Mol.Pharmacol., 67, 2005
|
|
7FC0
 
 | | Reconstitution of MbnABC complex from Rugamonas rubra ATCC-43154 (GroupIII) | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Chao, D, Zhaolin, L, Shoujie, L, Li, Z, Dan, Z, Ying, J, Wei, C. | | Deposit date: | 2021-07-13 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
