6BVY
 
 | | SFTI-HFRW-4 | | Descriptor: | Trypsin inhibitor 1 HFRW-4 | | Authors: | Schroeder, C.I, White, A. | | Deposit date: | 2017-12-14 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J. Med. Chem., 61, 2018
|
|
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6BVU
 
 | | SFTI-HFRW-1 | | Descriptor: | Trypsin inhibitor 1 HFRW-1 | | Authors: | Schroeder, C.I. | | Deposit date: | 2017-12-13 | | Release date: | 2018-12-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
7YOA
 
 | |
6JQ3
 
 | | Crystal Structure of H2-Kb in complex with a DPAGT1 mutant peptide | | Descriptor: | Beta-2-microglobulin, DPAGT1 mutant antigen SIIVFNLL, H-2 class I histocompatibility antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Yin, L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6JTN
 
 | | Crystal structure of HLA-C08 in complex with a tumor mut10m peptide | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Yin, L. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6K5M
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa (Rice) | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Liu, X.K, Guo, Y, Zhao, Y.C, Zeng, Z.X. | | Deposit date: | 2019-05-29 | | Release date: | 2020-06-03 | | Last modified: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6JBT
 
 | | Complex structure of toripalimab-Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Guo, L, Tan, S, Chai, Y, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy.
Mabs, 11, 2019
|
|
3CDE
 
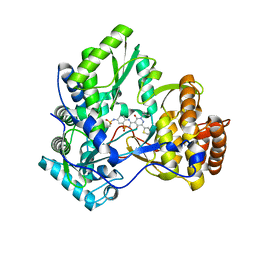 | | Crystal structure of HCV NS5B polymerase with a novel Pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-02-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 3: Further optimization of the 2-, 6-, and 7'-substituents and initial pharmacokinetic assessments.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6MJ2
 
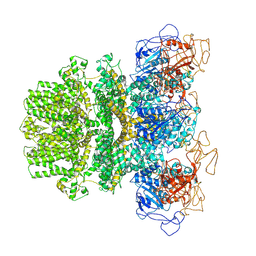 | | Human TRPM2 ion channel in a calcium- and ADPR-bound state | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-02 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
6MIX
 
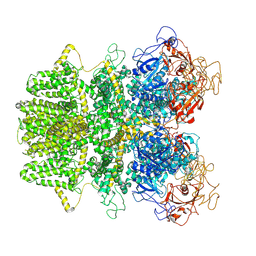 | | Human TRPM2 ion channel in apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
8GVJ
 
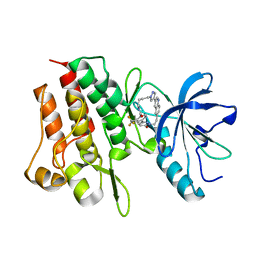 | | Crystal structure of cMET kinase domain bound by D6808 | | Descriptor: | (1^4Z,5^2E)-6^3-(trifluoromethyl)-5^1,5^6-dihydro-1^1H-8-aza-2(3,6)-quinolina-5(1,3)-pyridazina-1(4,1)-pyrazola-6(1,4)-benzenacyclododecaphane-5^6,7-dione, Hepatocyte growth factor receptor | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2022-09-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
6MIZ
 
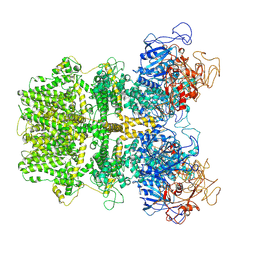 | |
8GZF
 
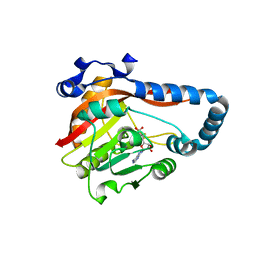 | | Crystal Structure of METTL9-SAH | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
8GZE
 
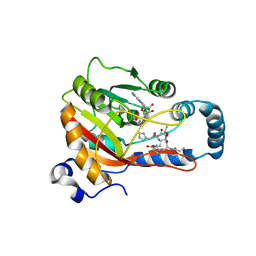 | | Crystal Structure of human METTL9-SAH-SLC39A7 peptide complex | | Descriptor: | Protein-L-histidine N-pros-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, Zinc transporter SLC39A7 | | Authors: | Zhao, W.T, Li, H.T. | | Deposit date: | 2022-09-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for protein histidine N1-specific methylation of the "His-x-His" motifs by METTL9.
Cell Insight, 2, 2023
|
|
6NW9
 
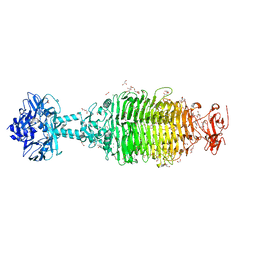 | | CRYSTAL STRUCTURE OF A TAILSPIKE PROTEIN 3 (TSP3, ORF212) FROM ESCHERICHIA COLI O157:H7 BACTERIOPHAGE CBA120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J.Y, Herzberg, O. | | Deposit date: | 2019-02-06 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3).
Sci Rep, 9, 2019
|
|
6JQ2
 
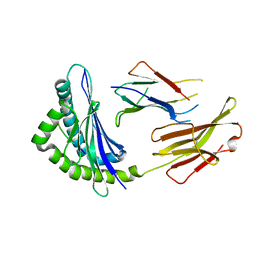 | | Crystal Structure of H2-Kb in complex with a DPAGT1 self-peptide | | Descriptor: | Beta-2-microglobulin, DPATG1 antigen SIIVFNLV, H-2 class I histocompatibility antigen, ... | | Authors: | Bai, P, Yin, L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6OBD
 
 | |
7MSD
 
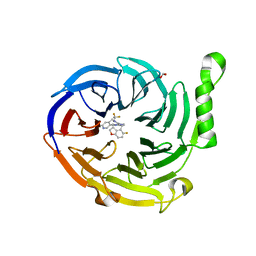 | | Structure of EED bound to EEDi-6068 | | Descriptor: | (9aP,12aR)-4-(2,2-difluoropropyl)-12-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,8,11,12a-pentaazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, FORMIC ACID, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7MSB
 
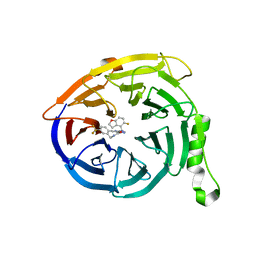 | | Structure of EED bound to EEDi-4259 | | Descriptor: | (9aM,12aS)-12-{[(5-fluoro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,11,12a-tetraazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7MBH
 
 | | Structure of Human Enolase 2 in complex with phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Gamma-enolase, ... | | Authors: | Leonard, P.G, Hicks, K.G, Rutter, J. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase.
Science, 379, 2023
|
|
5IMX
 
 | |
2JXW
 
 | |
7MRY
 
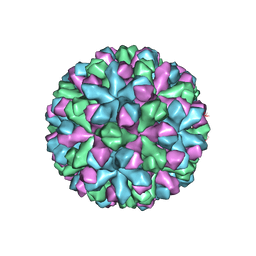 | | Norovirus T=3 GII.4 HOV VLP | | Descriptor: | VP1 | | Authors: | Salmen, W, Hu, L, Prasad, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the predominant GII.4 human norovirus capsid reveals novel stability and plasticity.
Nat Commun, 13, 2022
|
|
5DX4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-2-methoxybenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
