5NFS
 
 | |
5DUM
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody 65C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 65C6 Heavy Chain, 65C6 Light Chain, ... | | Authors: | Sun, J, Zuo, T, Wang, G, Zhou, P, Zhang, L, Wang, X. | | Deposit date: | 2015-09-19 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
5DUR
 
 | | Influenza A virus H5 hemagglutinin globular head in complex with antibody 100F4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Antibody 100F4, Hemagglutinin, ... | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6S24
 
 | | Crystal structure of the TgGalNAc-T3 in complex with UDP, manganese and the peptide 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-THR-GLY-ALA-GLY-ALA-GLY-ALA-GLY-THR-THR-PRO-GLY-PRO, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Narimatsu, Y, Companon, I, Kato, K, Hermosilla, P, Thureau, A, Ceballos-Laita, L, Coelho, H, Bernado, P, Marcelo, F, Hansen, L, Lostao, A, Corzana, F, Clausen, H, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3.
Nat.Chem.Biol., 16, 2020
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6WBX
 
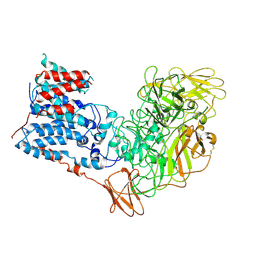 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WBY
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6X0O
 
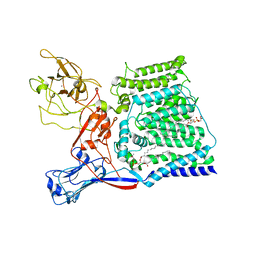 | | Single-Particle Cryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB | | Authors: | Tan, Y.Z, Rodrigues, J, Keener, J.E, Zheng, R.B, Brunton, R, Kloss, B, Giacometti, S.I, Rosario, A.L, Zhang, L, Niederweis, M, Clarke, O.B, Lowary, T.L, Marty, M.T, Archer, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis.
Nat Commun, 11, 2020
|
|
7O14
 
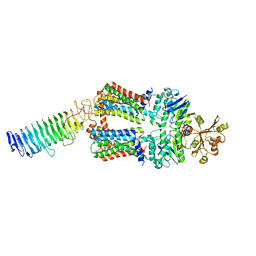 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O15
 
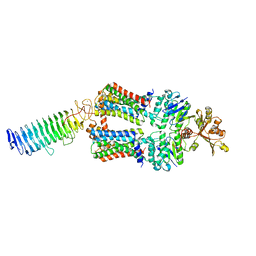 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O12
 
 | | ABC transporter NosDFY, AMPPNP-bound in GDN | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
6WN2
 
 | |
7O11
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O0Y
 
 | | ABC transporter NosDFY, nucleotide-free in GDN | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O16
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 3 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O10
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O17
 
 | | ABC transporter NosDFY E154Q, ATP-bound in lipid nanodisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
4GWG
 
 | |
8A4T
 
 | | crystal structures of diastereomer (S,S,S)-13b (13b-K) in complex with the SARS-CoV-2 Mpro | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
8A4Q
 
 | | crystal structures of diastereomer (R,S,S)-13b (13b-H) in complex with the SARS-CoV-2 Mpro. | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
8CTJ
 
 | | Cryo-EM structure of TMEM87A | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Transmembrane protein 87A | | Authors: | Hoel, C.M, Zhang, L, Brohawn, S.G. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-20 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.74 Å) | | Cite: | Structure of the GOLD-domain seven-transmembrane helix protein family member TMEM87A.
Elife, 11, 2022
|
|
