5FT2
 
 | | Sub-tomogram averaging of Lassa virus glycoprotein spike from virus- like particles at pH 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRE-GLYCOPROTEIN POLYPROTEIN GP COMPLEX | | Authors: | Li, S, Zhaoyang, S, Pryce, R, Parsy, M.L, Fehling, S.K, Schlie, K, Siebert, C.A, Garten, W, Bowden, T.A, Strecker, T, Huiskonen, J.T. | | Deposit date: | 2016-01-09 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Acidic Ph-Induced Conformations and Lamp1 Binding of the Lassa Virus Glycoprotein Spike.
Plos Pathog., 12, 2016
|
|
5FET
 
 | | Crystal Structure of PVX_084705 in presence of Compound 2 | | Descriptor: | 4-[7-[(dimethylamino)methyl]-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl]pyrimidin-2-amine, cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, Walker, J.R, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of PVX_084705 in presence of Compound 2
To be published
|
|
5FDU
 
 | | Crystal structure of the Metalnikowin I antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
5F83
 
 | | Imipenem complex of the GES-5 C69G mutant | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-12-09 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Role of the Conserved Disulfide Bridge in Class A Carbapenemases.
J.Biol.Chem., 291, 2016
|
|
5FFL
 
 | |
5F82
 
 | | Apo GES-5 C69G mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-12-08 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Role of the Conserved Disulfide Bridge in Class A Carbapenemases.
J.Biol.Chem., 291, 2016
|
|
5FDV
 
 | | Crystal structure of the Pyrrhocoricin antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
5FP0
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, N-cyclopentyl-2-[4-(trifluoromethyl)phenyl]-3H-benzimidazole-4-sulfonamide | | Authors: | Xue, Y, Olsson, T, Johansson, C.A, Oster, L, Beisel, H.G, Rohman, M, Karis, D, Backstrom, S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment Screening of Soluble Epoxide Hydrolase for Lead Generation-Structure-Based Hit Evaluation and Chemistry Exploration.
Chemmedchem, 11, 2016
|
|
5GAI
 
 | | Probabilistic Structural Models of Mature P22 Bacteriophage Portal, Hub, and Tailspike proteins | | Descriptor: | Peptidoglycan hydrolase gp4, Portal protein, Tail fiber protein | | Authors: | Pintilie, G, Chen, D.H, Haase-Pettingell, C.A, King, J.A, Chiu, W. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage.
Biophys.J., 110, 2016
|
|
5G5T
 
 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii in complex with guide DNA | | Descriptor: | ARGONAUTE, GUIDE DNA, MAGNESIUM ION, ... | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
5G5S
 
 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGONAUTE, MAGNESIUM ION | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
5HN1
 
 | | Crystal structure of Interleukin-37 | | Descriptor: | Interleukin-37, SULFATE ION | | Authors: | Ellisdon, A.M, Nold-Petry, C.A, Nold, M.F, Whisstock, J.C. | | Deposit date: | 2016-01-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Homodimerization attenuates the anti-inflammatory activity of interleukin-37.
Sci Immunol, 2, 2017
|
|
5HGR
 
 | | Structure of Anabaena (Nostoc) sp. PCC 7120 Orange Carotenoid Protein binding canthaxanthin | | Descriptor: | Orange Carotenoid Protein (OCP), beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Leverenz, R.L, Kerfeld, C.A. | | Deposit date: | 2016-01-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Different Functions of the Paralogs to the N-Terminal Domain of the Orange Carotenoid Protein in the Cyanobacterium Anabaena sp. PCC 7120.
Plant Physiol., 171, 2016
|
|
5I08
 
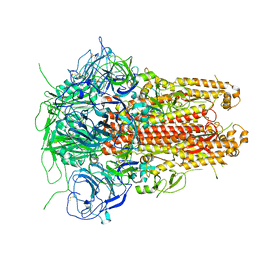 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
5I35
 
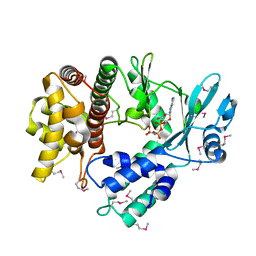 | | Structure of the Human mitochondrial kinase COQ8A R611K with AMPPNP (Cerebellar Ataxia and Ubiquinone Deficiency Through Loss of Unorthodox Kinase Activity) | | Descriptor: | Atypical kinase ADCK3, mitochondrial, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bingman, C.A, Stefely, J.A, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity.
Mol.Cell, 63, 2016
|
|
3ZK7
 
 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
3ZKA
 
 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA D280N IN THE METAL-BOUND, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
3ZK8
 
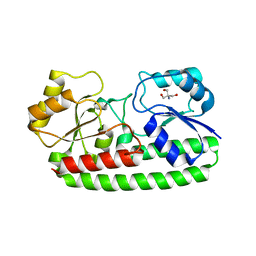 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA E205Q IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
3ZK9
 
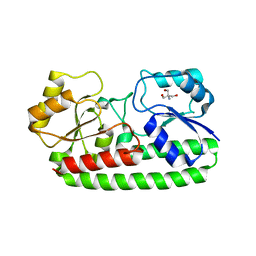 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA D280N IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
1F9N
 
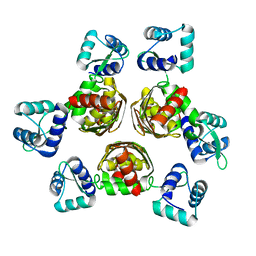 | | CRYSTAL STRUCTURE OF AHRC, THE ARGININE REPRESSOR/ACTIVATOR PROTEIN FROM BACILLUS SUBTILIS | | Descriptor: | ARGININE REPRESSOR/ACTIVATOR PROTEIN | | Authors: | Dennis, C.A, Glykos, N.M, Parsons, M.R, Phillips, S.E.V. | | Deposit date: | 2000-07-11 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4NST
 
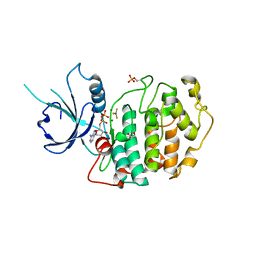 | | Crystal structure of human Cdk12/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Boesken, C.A, Farnung, L, Anand, K, Geyer, M. | | Deposit date: | 2013-11-29 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure and substrate specificity of human Cdk12/Cyclin K.
Nat Commun, 5, 2014
|
|
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
4OM9
 
 | | X-Ray Crystal Structure of the passenger domain of Plasmid encoded toxin, an Autrotansporter Enterotoxin from enteroaggregative Escherichia coli (EAEC) | | Descriptor: | Serine protease pet | | Authors: | Meza-Aguilar, J.D, Fromme, P, Torres-Larios, A, Mendoza-Hernandez, G, Hernandez-Chinas, U, Arreguin-Espinosa de Los Monteros, R.A, Eslava-Campos, C.A, Fromme, R. | | Deposit date: | 2014-01-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the passenger domain of plasmid encoded toxin(Pet), an autotransporter enterotoxin from enteroaggregative Escherichia coli (EAEC).
Biochem.Biophys.Res.Commun., 445, 2014
|
|
2F00
 
 | | Escherichia coli MurC | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Deva, T, Baker, E.N, Squire, C.J, Smith, C.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Escherichia coliUDP-N-acetylmuramoyl:L-alanine ligase (MurC).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2FNS
 
 | | Crystal structure of wild-type inactive (D25N) HIV-1 protease complexed with wild-type HIV-1 NC-p1 substrate. | | Descriptor: | ACETATE ION, NC-P1 SUBSTRATE PEPTIDE, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, Schiffer, C.A. | | Deposit date: | 2006-01-11 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate.
J.Virol., 80, 2006
|
|
