2KPR
 
 | |
2OE8
 
 | | 1.8 A X-ray crystal structure of Apramycin complex with RNA fragment GGGCGUCGCUAGUACC/CGGUACUAAAAGUCGCC containing the human ribosomal decoding A site: RNA construct with 5'-overhang | | Descriptor: | APRAMYCIN, RNA (5'-R(*CP*GP*GP*UP*AP*CP*UP*AP*AP*AP*AP*GP*UP*CP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*C)-3') | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
2O3M
 
 | | Monomeric G-DNA tetraplex from human C-kit promoter | | Descriptor: | 5'-D(*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*CP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Burge, S, Neidle, S, Patel, D.J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter.
J.Am.Chem.Soc., 129, 2007
|
|
2OE5
 
 | | 1.5 A X-ray crystal structure of Apramycin complex with RNA fragment GGCGUCGCUAGUACCG/GGUACUAAAAGUCGCCC containing the human ribosomal decoding A site: RNA construct with 3'-overhang | | Descriptor: | APRAMYCIN, MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*CP*G)-3'), ... | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
7W27
 
 | | Crystal structure of BEND3-BEN4-DNA complex | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*CP*GP*TP*GP*GP*GP*TP*C)-3') | | Authors: | Zheng, L, Ren, A. | | Deposit date: | 2021-11-22 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins.
Genes Dev., 36, 2022
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
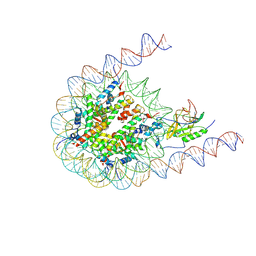 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
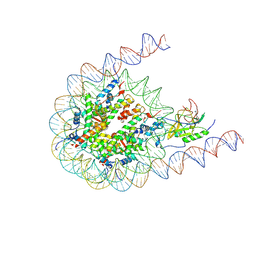 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
5XP8
 
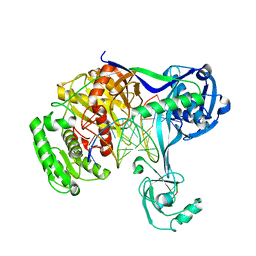 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 4A5 on the guide strand | | Descriptor: | DNA (5'-D(*AP*GP*T)-3'), DNA (5'-D(*CP*AP*AP*CP*C*AP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*AP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*T)-3'), ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, J.D, Wang, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Nucleic Acids Res., 45, 2017
|
|
5XQ2
 
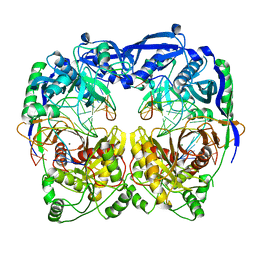 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 5A6 on the guide strand | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(*AP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*AP*GP*AP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*T)-3'), ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, D, Wang, Y. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes
Nucleic Acids Res., 45, 2017
|
|
5XPA
 
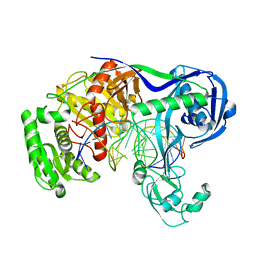 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 9'U10' on the target strand | | Descriptor: | DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), MAGNESIUM ION, RNA (5'-R(P*AP*UP*AP*CP*AP*AP*CP*CP*GP*UP*UP*CP*UP*AP*CP*UP*CP*CP*G)-3'), ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, J.D, Wang, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Nucleic Acids Res., 45, 2017
|
|
5XPG
 
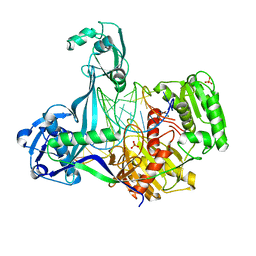 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 6'U7' on the target strand | | Descriptor: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*A P*GP*T)-3', 5'-R(*UP*AP*U*AP*CP*AP*AP*CP*CP*UP*AP*CP*AP*UP*AP*CP*CP*UP*CP* G)-3', MAGNESIUM ION, ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, J.D, Wang, Y. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Nucleic Acids Res., 45, 2017
|
|
3F2W
 
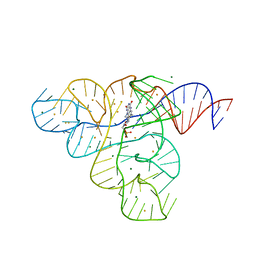 | |
3NCU
 
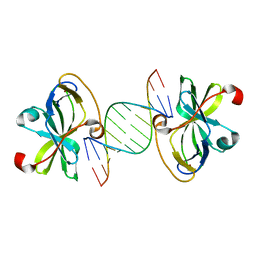 | |
7VKI
 
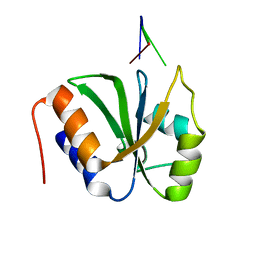 | | ESRP1 qRRM2 in complex with 12mer-RNA | | Descriptor: | Epithelial splicing regulatory protein 1, RNA (12-mer) | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ESRP1 controls biogenesis and function of a large abundant multiexon circRNA.
Nucleic Acids Res., 2023
|
|
6DBP
 
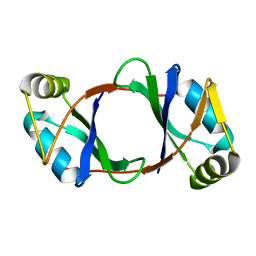 | |
2GDI
 
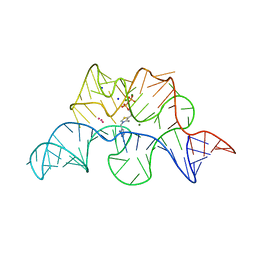 | |
6JQ5
 
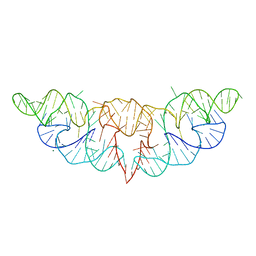 | | The structure of Hatchet Ribozyme | | Descriptor: | MAGNESIUM ION, RNA (82-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JQ6
 
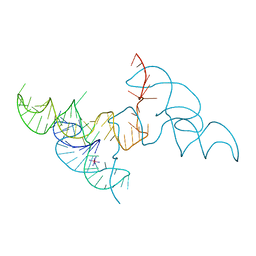 | | Hatchet Ribozyme Structure soaking with Ir(NH3)6+ | | Descriptor: | IRIDIUM HEXAMMINE ION, RNA (81-MER) | | Authors: | Ren, A, Zheng, L. | | Deposit date: | 2019-03-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Hatchet ribozyme structure and implications for cleavage mechanism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4X0G
 
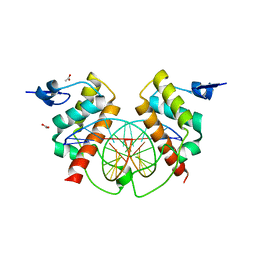 | | Structure of Bsg25A binding with DNA | | Descriptor: | ACETATE ION, Blastoderm-specific gene 25A, DNA (5'-D(*GP*TP*TP*CP*CP*AP*AP*TP*TP*GP*GP*AP*A)-3') | | Authors: | Ren, A. | | Deposit date: | 2014-11-21 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2065 Å) | | Cite: | Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family.
Genes Dev., 29, 2015
|
|
4KPY
 
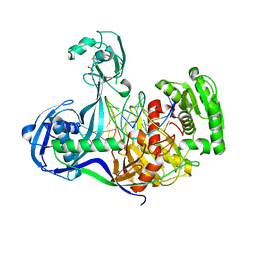 | | DNA binding protein and DNA complex structure | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3F2X
 
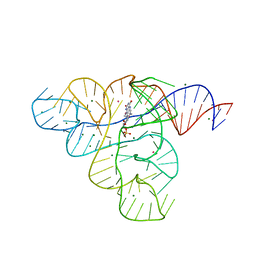 | |
3F4E
 
 | |
5HH7
 
 | |
