8HP7
 
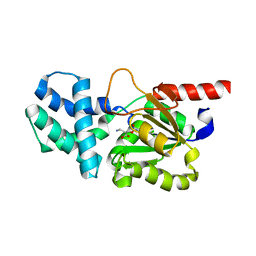 | | Crystal structure of (S)-2-haloacid dehalogenase K152A mutant trapped with (2R)-4-amino-2-hydroxybutanoic acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
6JKF
 
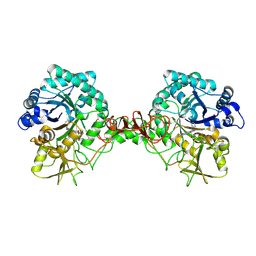 | | Crystal structure of Serratia marcescens Chitinase B complexed with compound 2-8-s2 | | Descriptor: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitinase | | Authors: | Yang, Q, Jiang, X. | | Deposit date: | 2019-02-28 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
4WTR
 
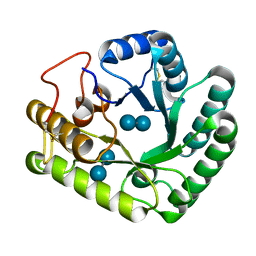 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTP
 
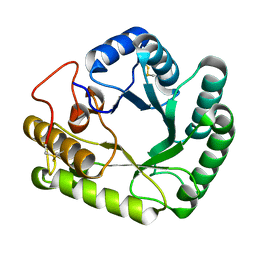 | | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei | | Descriptor: | 1,2-ETHANEDIOL, beta-1,3-glucanosyltransferase | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTS
 
 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WY5
 
 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | Esterase, SULFATE ION | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
4WY8
 
 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | esterase | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
6QEM
 
 | | E. coli DnaBC complex bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein DnaC, MAGNESIUM ION, ... | | Authors: | Arias-Palomo, E, Puri, N, O'Shea Murray, V.L, Yan, Q, Berger, J.M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.
Mol.Cell, 74, 2019
|
|
6QEL
 
 | | E. coli DnaBC apo complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein dnaC, MAGNESIUM ION, ... | | Authors: | Arias-Palomo, E, Puri, N, O'Shea Murray, V.L, Yan, Q, Berger, J.M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.
Mol.Cell, 74, 2019
|
|
5H9X
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | Descriptor: | beta-1,3-glucanase | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5H9Y
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | Descriptor: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5JVV
 
 | | Crystal structure and characterization an elongating GH family 16 beta-1,3-glucosyltransferase | | Descriptor: | beta-1,3-glucosyltransferase | | Authors: | Qin, Z, Yan, Q, Yang, S, Jiang, Z. | | Deposit date: | 2016-05-11 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Catalytic Mechanism of a Novel Glycoside Hydrolase Family 16 "Elongating" beta-Transglycosylase
J. Biol. Chem., 292, 2017
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2QE3
 
 | | Crystal structure of human tl1a extracellular domain | | Descriptor: | CHLORIDE ION, TNF superfamily ligand TL1A | | Authors: | Zhan, C, Yan, Q, Patskovsky, Y, Shi, W, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-06-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
2RDM
 
 | | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Yan, Q, Zhan, C, Toro, R, Meyer, A.J, Gilmore, M, Hu, S, Groshong, C, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419.
To be Published
|
|
2RJK
 
 | | Crystal Structure of Human TL1A Extracellular Domain C95S Mutant | | Descriptor: | TNF superfamily ligand TL1A | | Authors: | Zhan, C, Yan, Q, Patskovsky, Y, Shi, W, Ramagopal, U.A, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-10-15 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
2RJL
 
 | | Crystal structure of human TL1A extracellular domain C95S/C135S mutant | | Descriptor: | TNF superfamily ligand TL1A | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Shi, W, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-10-15 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
5XBZ
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
5XC2
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminarihexaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Qin, Z, Yang, S, Peng, Z, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
3X21
 
 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
3K51
 
 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3MI8
 
 | | The structure of TL1A-DCR3 COMPLEX | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 15, SECRETED FORM, Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-09 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3RP1
 
 | | Crystal structure of Human LAIR-1 in C2 space group | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1 | | Authors: | Sampathkumar, P, Ramagopal, U.A, Yan, Q, Toro, R, Nathenson, S, Bonanno, J, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Human LAIR-1 in C2 space group
To be Published
|
|
3MHD
 
 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
8HE4
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
