8B2B
 
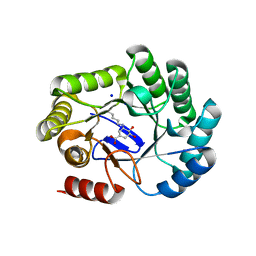 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, SODIUM ION | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
4UIO
 
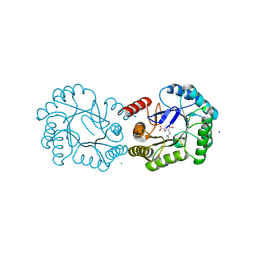 | | Structure of the Salmonella typhi Type I Dehydroquinase covalently inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1~{R},3~{R},4~{S},5~{R})-3-methyl-1,3,4,5-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Tizon, L, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Chemical Modification of a Dehydratase Enzyme Involved in Bacterial Virulence by an Ammonium Derivative: Evidence of its Active Site Covalent Adduct.
J.Am.Chem.Soc., 137, 2015
|
|
4UE0
 
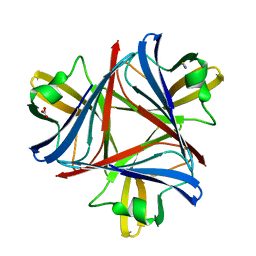 | |
6SFE
 
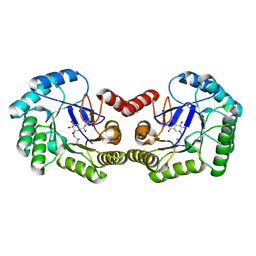 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
1OCY
 
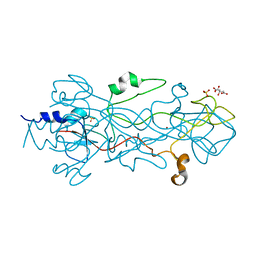 | | Structure of the receptor-binding domain of the bacteriophage T4 short tail fibre | | Descriptor: | BACTERIOPHAGE T4 SHORT TAIL FIBRE, CITRIC ACID, SULFATE ION, ... | | Authors: | Thomassen, E, Gielen, G, Schuetz, M, Miller, S, van Raaij, M.J. | | Deposit date: | 2003-02-11 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Receptor-Binding Domain of the Bacteriophage T4 Short Tail Fibre Reveals a Knitted Trimeric Metal-Binding Fold
J.Mol.Biol., 331, 2003
|
|
1PDI
 
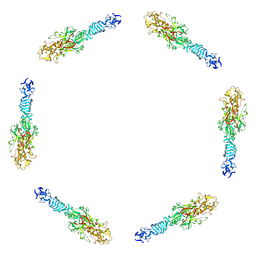 | | Fitting of the C-terminal part of the short tail fibers into the cryo-EM reconstruction of T4 baseplate | | Descriptor: | Short tail fiber protein | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1EFR
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH THE PEPTIDE ANTIBIOTIC EFRAPEPTIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT ALPHA, BOVINE MITOCHONDRIAL F1-ATPASE SUBUNIT BETA, ... | | Authors: | Abrahams, J.P, Buchanan, S.K, Van Raaij, M.J, Fearnley, I.M, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-24 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of Bovine F1-ATPase Complexed with the Peptide Antibiotic Efrapeptin.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5FLD
 
 | |
5FWZ
 
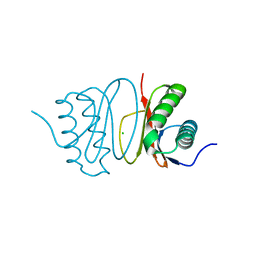 | | Fasciola hepatica calcium binding protein FhCaBP2: Structure of the dynein light chain-like domain. P41212 mercury derivative. | | Descriptor: | CALCIUM BINDING PROTEIN, CHLORIDE ION, MERCURY (II) ION | | Authors: | Nguyen, T.H, Thomas, C.M, Timson, D.J, van Raaij, M.J. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fasciola hepatica calcium-binding protein FhCaBP2: structure of the dynein light chain-like domain.
Parasitol. Res., 115, 2016
|
|
5G5O
 
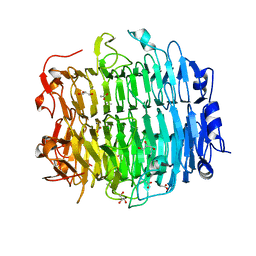 | | Structure of the snake adenovirus 1 hexon-interlacing LH3 protein, native | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nguyen, T.H, Singh, A.K, Albala-Perez, B, van Raaij, M.J. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid.
Structure, 25, 2017
|
|
5FJL
 
 | |
5G5N
 
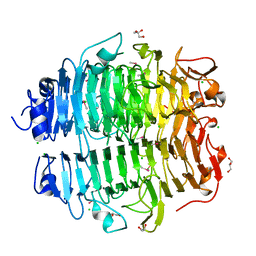 | | Structure of the snake adenovirus 1 hexon-interlacing LH3 protein, methylmercury chloride derivative | | Descriptor: | CHLORIDE ION, GLYCEROL, LH3 HEXON-INTERLACING CAPSID PROTEIN, ... | | Authors: | Nguyen, T.H, Singh, A.K, Albala-Perez, B, van Raaij, M.J. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid.
Structure, 25, 2017
|
|
6SFH
 
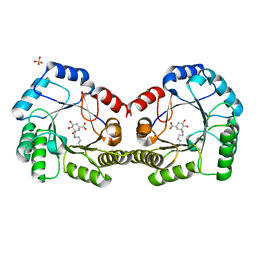 | | CRYSTAL STRUCTURE OF DHQ1 FROM Staphylococcus aureus COVALENTLY MODIFIED BY LIGAND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase, LITHIUM ION, ... | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
6H5J
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 4 | | Descriptor: | (3~{R})-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Otero, J.M, Maneiro, M, Lence, E, Sanz-Gaitero, M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
6H5C
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 1 | | Descriptor: | (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
6H5D
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 2 | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 9, 2019
|
|
6H5G
 
 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 3 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
6SFG
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 9 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
2WSU
 
 | | Galectin domain of porcine adenovirus type 4 NADC-1 isolate fibre | | Descriptor: | GLYCEROL, NITRATE ION, PUTATIVE FIBER PROTEIN | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2WST
 
 | | Head domain of porcine adenovirus type 4 NADC-1 isolate fibre | | Descriptor: | PUTATIVE FIBER PROTEIN | | Authors: | Guardado-Calvo, P, Munoz, E.M, Llamas-Saiz, A.L, Fox, G.C, Glasgow, J.N, van Raaij, M.J. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
J. Virol., 84, 2010
|
|
2XGF
 
 | | Structure of the bacteriophage T4 long tail fibre needle-shaped receptor-binding tip | | Descriptor: | CARBONATE ION, FE (II) ION, LONG TAIL FIBER PROTEIN P37 | | Authors: | Bartual, S.G, Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Kahn, R, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2010-06-03 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacteriophage T4 long tail fiber receptor-binding tip.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
5M9F
 
 | |
5NBH
 
 | |
5NC1
 
 | |
5N83
 
 | |
